7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
7D0X
 
 | |
8X4F
 
 | |
7WRA
 
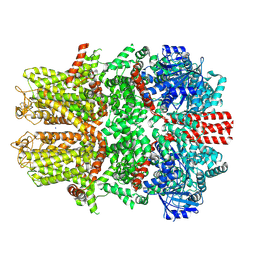 | | Mouse TRPM8 in LMNG in ligand-free state | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
6PVP
 
 | |
6S3T
 
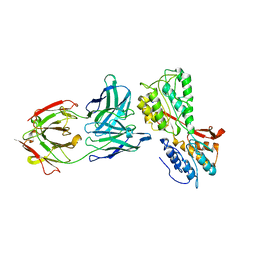 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | 46 kDa surface antigen, Immunoglobulin heavy chain, Immunoglobulin light chain, ... | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2GVH
 
 | |
1VC5
 
 | | Crystal Structure of the Wild Type Hepatitis Delta Virus Gemonic Ribozyme Precursor, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, SODIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
3US9
 
 | | Crystal Structure of the NCX1 Intracellular Tandem Calcium Binding Domains(CBD12) | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Giladi, M, Sasson, Y, Hirsch, J.A, Khananshvili, D. | | Deposit date: | 2011-11-23 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation.
Plos One, 7, 2012
|
|
1G5U
 
 | | LATEX PROFILIN HEVB8 | | Descriptor: | PROFILIN, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Ganglberger, E, Breiteneder, H, Almo, S.C. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Comparative Structural Analysis of Allergen Profilins HEVB8 and BETV2
To be Published
|
|
3G40
 
 | |
8ULF
 
 | |
4V9F
 
 | |
6NLH
 
 | | Structure of human triose phosphate isomerase R189A | | Descriptor: | BROMIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Richards, K.R, Roland, B.P, Palladino, M.J, VanDemark, A.P. | | Deposit date: | 2019-01-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Missense variant in TPI1 (Arg189Gln) causes neurologic deficits through structural changes in the triosephosphate isomerase catalytic site and reduced enzyme levels in vivo.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
6TBH
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-[ethyl(methyl)amino]-~{N}-[6-[[(1~{S},2~{R},3~{S},4~{R})-2-(hydroxymethyl)-3,4-bis(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6TBG
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[[(1~{R},2~{R},3~{S},4~{S},5~{S})-2-(hydroxymethyl)-3,4,5-tris(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6Z9F
 
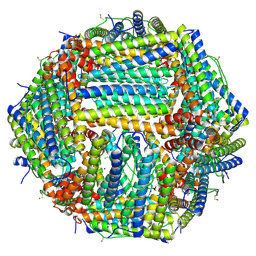 | | 1.56 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.56 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6TBJ
 
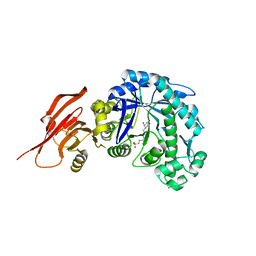 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[(2~{S},3~{R},4~{S},5~{R})-3-(hydroxymethyl)-4,5-bis(oxidanyl)piperidin-2-yl]hexyl]naphthalene-1-sulfonamide, Beta-galactosidase, putative, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
6Z9E
 
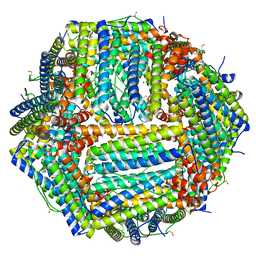 | | 1.55 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6Z6U
 
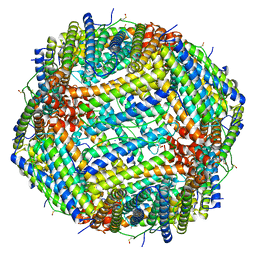 | | 1.25 A structure of human apoferritin obtained from Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-05-29 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.25 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6DWD
 
 | | SAMHD1 Bound to Clofarabine-TP in the Catalytic Pocket and Allosteric Pocket | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, 9-{2-deoxy-2-fluoro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-2-me thyl-9H-purin-6-amine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W1F
 
 | | Crystal structure of Ni(II)- and Ca(II)-bound human calprotectin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Protein S100-A8, ... | | Authors: | Nakashige, T.G, Drennan, C.L, Nolan, E.M. | | Deposit date: | 2017-06-03 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nickel Sequestration by the Host-Defense Protein Human Calprotectin.
J. Am. Chem. Soc., 139, 2017
|
|
3FFC
 
 | | Crystal Structure of CF34 TCR in complex with HLA-B8/FLR | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CF34 alpha chain, ... | | Authors: | Gras, S, Burrows, S.R, Kjer-Nielsen, L, Clements, C.S, Liu, Y.C, Sullivan, L.C, Brooks, A.G, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shaping of T cell receptor recognition by self-tolerance.
Immunity, 30, 2009
|
|
1YI2
 
 | | Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-11 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YIT
 
 | | Crystal Structure Of Virginiamycin M and S Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-04-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
