1GGH
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, HIS128ALA VARIANT. | | Descriptor: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGF
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, VARIANT HIS128ASN, COMPLEX WITH HYDROGEN PEROXIDE. | | Descriptor: | CATALASE HPII, HYDROGEN PEROXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1F5Y
 
 | | NMR STRUCTURE OF A CONCATEMER OF THE FIRST AND SECOND LIGAND-BINDING MODULES OF THE HUMAN LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Atkins, A.R, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-06-18 | | Release date: | 2000-08-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor.
Protein Sci., 9, 2000
|
|
1F6M
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THIOREDOXIN REDUCTASE, THIOREDOXIN, AND THE NADP+ ANALOG, AADP+ | | Descriptor: | 3-AMINOPYRIDINE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN 1, ... | | Authors: | Lennon, B.W, Williams Jr, C.H, Ludwig, M.L. | | Deposit date: | 2000-06-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase.
Science, 289, 2000
|
|
1F61
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE LYASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ISOCITRATE LYASE, MAGNESIUM ION | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K.H, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-19 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1E5G
 
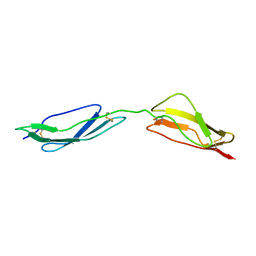 | | Solution structure of central CP module pair of a pox virus complement inhibitor | | Descriptor: | COMPLEMENT CONTROL PROTEIN C3 | | Authors: | Henderson, C.E, Bromek, K, Mullin, N.P, Smith, B.O, Uhrin, D, Barlow, P.N. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-31 | | Last modified: | 2013-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Central Ccp Module Pair of a Poxvirus Complement Control Protein
J.Mol.Biol., 307, 2001
|
|
1GCV
 
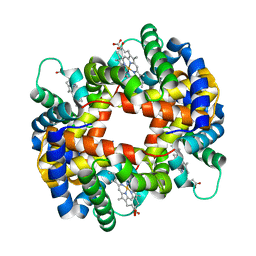 | | DEOXY FORM HEMOGLOBIN FROM MUSTELUS GRISEUS | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Naoi, Y, Chong, K.T, Yoshimatsu, K, Miyazaki, G, Tame, J.R.H, Park, S.Y, Adachi, S.I, Morimoto, H. | | Deposit date: | 2000-08-08 | | Release date: | 2000-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The functional similarity and structural diversity of human and cartilaginous fish hemoglobins.
J.Mol.Biol., 307, 2001
|
|
1QB3
 
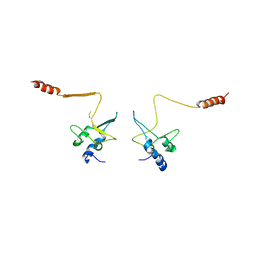 | | CRYSTAL STRUCTURE OF THE CELL CYCLE REGULATORY PROTEIN CKS1 | | Descriptor: | CYCLIN-DEPENDENT KINASES REGULATORY SUBUNIT | | Authors: | Bourne, Y, Watson, M.H, Arvai, A.S, Bernstein, S.L, Reed, S.I, Tainer, J.A. | | Deposit date: | 1999-04-30 | | Release date: | 2000-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mutational analysis of the Saccharomyces cerevisiae cell cycle regulatory protein Cks1: implications for domain swapping, anion binding and protein interactions.
Structure Fold.Des., 8, 2000
|
|
1F6Y
 
 | |
1EKU
 
 | | CRYSTAL STRUCTURE OF A BIOLOGICALLY ACTIVE SINGLE CHAIN MUTANT OF HUMAN IFN-GAMMA | | Descriptor: | Interferon gamma, SULFATE ION | | Authors: | Landar, A, Curry, B, Parker, M.H, DiGiacomo, R, Indelicato, S.R, Walter, M.R. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, characterization, and structure of a biologically active single-chain mutant of human IFN-gamma.
J.Mol.Biol., 299, 2000
|
|
1F0Y
 
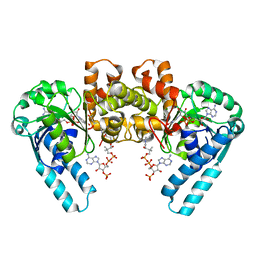 | | L-3-HYDROXYACYL-COA DEHYDROGENASE COMPLEXED WITH ACETOACETYL-COA AND NAD+ | | Descriptor: | ACETOACETYL-COENZYME A, L-3-HYDROXYACYL-COA DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequestration of the active site by interdomain shifting. Crystallographic and spectroscopic evidence for distinct conformations of L-3-hydroxyacyl-CoA dehydrogenase.
J.Biol.Chem., 275, 2000
|
|
1F1T
 
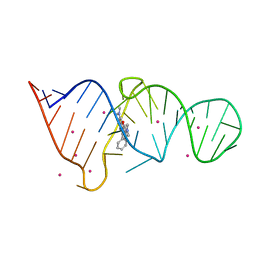 | | CRYSTAL STRUCTURE OF THE MALACHITE GREEN APTAMER COMPLEXED WITH TETRAMETHYL-ROSAMINE | | Descriptor: | MALACHITE GREEN APTAMER RNA, N,N'-TETRAMETHYL-ROSAMINE, STRONTIUM ION | | Authors: | Baugh, C, Grate, D, Wilson, C. | | Deposit date: | 2000-05-19 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A crystal structure of the malachite green aptamer.
J.Mol.Biol., 301, 2000
|
|
1FLO
 
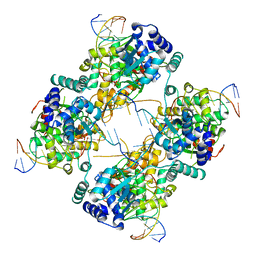 | | FLP Recombinase-Holliday Junction Complex I | | Descriptor: | FLP RECOMBINASE, PHOSPHONIC ACID, SYMMETRIZED FRT DNA SITES | | Authors: | Chen, Y, Narendra, U, Iype, L.E, Cox, M.M, Rice, P.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping.
Mol.Cell, 6, 2000
|
|
1DEB
 
 | |
1DIX
 
 | | CRYSTAL STRUCTURE OF RNASE LE | | Descriptor: | EXTRACELLULAR RIBONUCLEASE LE | | Authors: | Tanaka, N, Nakamura, K.T. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a plant ribonuclease, RNase LE.
J.Mol.Biol., 298, 2000
|
|
1EYQ
 
 | | Chalcone isomerase and naringenin | | Descriptor: | CHALCONE-FLAVONONE ISOMERASE 1, NARINGENIN, SULFATE ION | | Authors: | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-05-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
1EIW
 
 | |
1EFH
 
 | |
1ET1
 
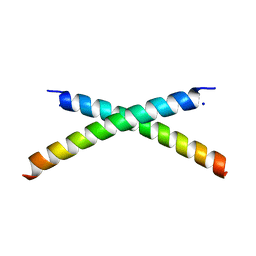 | | CRYSTAL STRUCTURE OF HUMAN PARATHYROID HORMONE 1-34 AT 0.9 A RESOLUTION | | Descriptor: | PARATHYROID HORMONE, SODIUM ION | | Authors: | Jin, L, Briggs, S.L, Chandrasekhar, S, Chirgadze, N.Y, Clawson, D.K, Schevitz, R.W, Smiley, D.L, Tashjian, A.H, Zhang, F. | | Deposit date: | 2000-04-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal structure of human parathyroid hormone 1-34 at 0.9-A resolution.
J.Biol.Chem., 275, 2000
|
|
1EYP
 
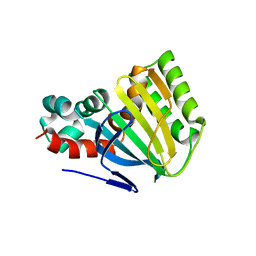 | | CHALCONE ISOMERASE | | Descriptor: | CHALCONE-FLAVONONE ISOMERASE 1 | | Authors: | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-05-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
1F2L
 
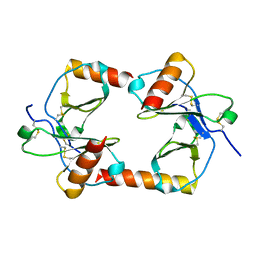 | |
1EZW
 
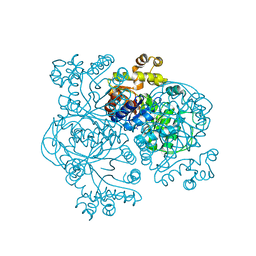 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1F07
 
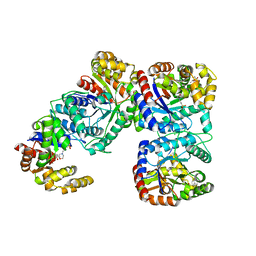 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1F0C
 
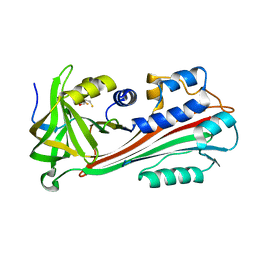 | | STRUCTURE OF THE VIRAL SERPIN CRMA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ICE INHIBITOR | | Authors: | Renatus, M, Zhou, Q, Stennicke, H.R, Snipas, S.J, Turk, D, Bankston, L.A, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the apoptotic suppressor CrmA in its cleaved form.
Structure Fold.Des., 8, 2000
|
|
1DVM
 
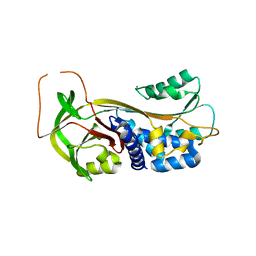 | | ACTIVE FORM OF HUMAN PAI-1 | | Descriptor: | CHLORIDE ION, PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Stout, T.J, Graham, H, Buckley, D.I, Matthews, D.J. | | Deposit date: | 2000-01-21 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of active and latent PAI-1: a possible stabilizing role for chloride ions.
Biochemistry, 39, 2000
|
|
