4P81
 
 | | Structure of ancestral PyrR protein (AncORANGEPyrR) | | Descriptor: | Ancestral PyrR protein (Orange), GLYCEROL, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
1GQY
 
 | | MURC - CRYSTAL STRUCTURE OF THE ENZYME FROM HAEMOPHILUS INFLUENZAE COMPLEXED WITH AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE | | Authors: | Skarzynski, T, Cleasby, A, Domenici, E, Gevi, M, Shaw, J. | | Deposit date: | 2001-12-06 | | Release date: | 2003-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Udp-N-Acetylmuramate-L-Alanine Ligase (Murc) from Haemophilus Influenzae
To be Published
|
|
1A1A
 
 | |
1A08
 
 | |
4P3K
 
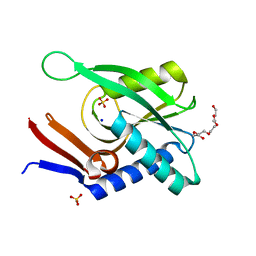 | | Structure of ancestral PyrR protein (PLUMPyrR) | | Descriptor: | Ancestral PyrR protein (Plum), PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
2ESK
 
 | |
3LEN
 
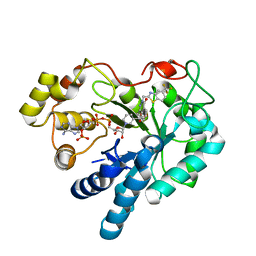 | | Human Aldose Reductase mutant T113S complexed with Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-15 | | Release date: | 2011-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
6MV6
 
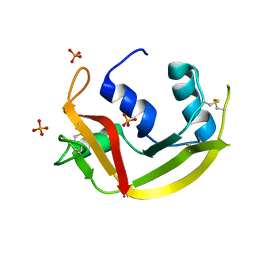 | | Crystal structure of RNAse 6 | | Descriptor: | PHOSPHATE ION, Ribonuclease K6 | | Authors: | Couture, J.-F, Doucet, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
1A09
 
 | |
6MW8
 
 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Manganese | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
1A1E
 
 | |
5L23
 
 | |
5F3K
 
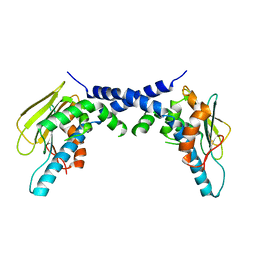 | | X-Ray Crystallographic Structure of hTrap1 N-terminal Domain-apo | | Descriptor: | Heat shock protein 75 kDa, mitochondrial | | Authors: | Sung, N, Lee, J, Kim, J, Chang, C, Joachimiak, A, Lee, S, Tsai, F.T.F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5INI
 
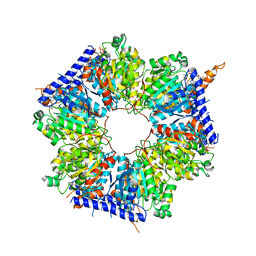 | | Structural basis for acyl-CoA carboxylase-mediated assembly of unusual polyketide synthase extender units incorporated into the stambomycin antibiotics | | Descriptor: | HEXANOYL-COENZYME A, Putative carboxyl transferase | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Withall, D.M, Song, L, Milligan, J.C, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
1A1B
 
 | |
3MC5
 
 | |
2VTD
 
 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2008-05-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
4AC8
 
 | | R2-like ligand binding Mn-Fe oxidase from M. tuberculosis with an organized C-terminal helix | | Descriptor: | CALCIUM ION, FE (III) ION, GLYCEROL, ... | | Authors: | Andersson, C.S, Berthold, C.L, Hogbom, M. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Dynamic C-Terminal Segment in the Mycobacterium Tuberculosis Mn/Fe R2Lox Protein Can Adopt a Helical Structure with Possible Functional Consequences.
Chem.Biodivers., 9, 2012
|
|
1A1C
 
 | |
6PTY
 
 | | Soluble model of human CuA (Tt3Lh) | | Descriptor: | Cytochrome c oxidase subunit 2, DINUCLEAR COPPER ION, GLYCEROL, ... | | Authors: | Giannini, E, Lisa, M.N, Morgada, M.N, Alzari, P.M, Vila, A.J. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unexpected electron spin density on the axial methionine ligand in CuAsuggests its involvement in electron pathways.
Chem.Commun.(Camb.), 56, 2020
|
|
1A07
 
 | |
4P82
 
 | | Structure of PyrR protein from Bacillus subtilis | | Descriptor: | Bifunctional protein PyrR, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
3E2B
 
 | | Crystal structure of Dynein Light chain LC8 in complex with a peptide derived from Swallow | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Barbar, E, Karplus, P.A. | | Deposit date: | 2008-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interplay of ligand binding and quaternary structure in the diverse interactions of dynein light chain LC8.
J.Mol.Biol., 384, 2008
|
|
4RVK
 
 | | CHK1 kinase domain with diazacarbazole compound 8: N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide | | Descriptor: | N-[3-(6-cyano-9H-pyrrolo[2,3-b:5,4-c']dipyridin-3-yl)phenyl]acetamide, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
1GQQ
 
 | |
