3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | Descriptor: | Computational design, MID1-apo1 | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
7XFA
 
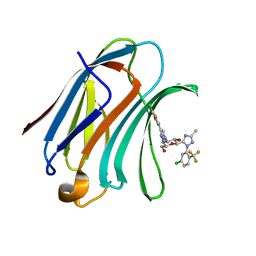 | | Structure of human Galectin-3 CRD in complex with monosaccharide inhibitor | | Descriptor: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[2-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Shukla, J, Raman, S, Ghosh, K. | | Deposit date: | 2022-04-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Identification of Monosaccharide Derivatives as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3.
J.Med.Chem., 65, 2022
|
|
5NFK
 
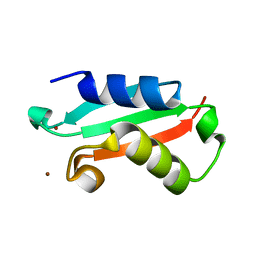 | |
8V56
 
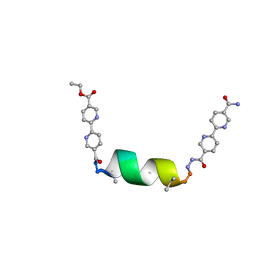 | | UIC-1 mutant - UIC-1-B5W | | Descriptor: | UIC-1-B5W | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-11-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
6SG6
 
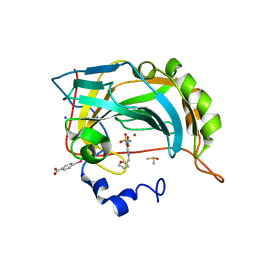 | |
3JU4
 
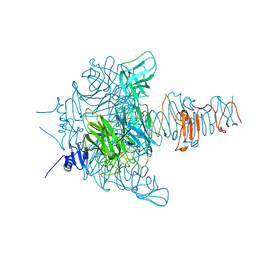 | | Crystal Structure Analysis of EndosialidaseNF at 0.98 A Resolution | | Descriptor: | CHLORIDE ION, Endo-N-acetylneuraminidase, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Neuman, P, Gerardy-Schahn, R, Sheldrick, G.M, Ficner, R. | | Deposit date: | 2009-09-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure analysis of endosialidase NF at 0.98 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3GGI
 
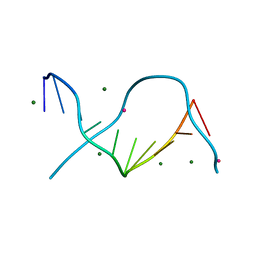 | | Locating monovalent cations in one turn of G/C rich B-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G) -3', MAGNESIUM ION, THALLIUM (I) ION | | Authors: | Maehigashi, T, Moulaei, T, Watkins, D, Komeda, S, Williams, L.D. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Locating monovalent cations in one turn of G/C rich B-DNA
To be Published
|
|
3JUD
 
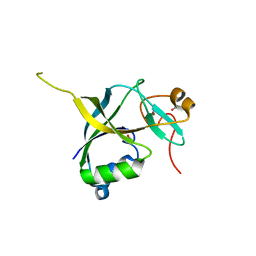 | | Human gamma-glutamylamine cyclotransferase, E82Q mutant | | Descriptor: | AIG2-like domain-containing protein 1, NITRATE ION | | Authors: | Oakley, A.J. | | Deposit date: | 2009-09-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Identification and characterization of {gamma}-glutamylamine cyclotransferase: An enzyme responsible for {gamma}-glutamyl-{epsilon}-lysine catabolism
J.Biol.Chem., 285, 2010
|
|
5SY4
 
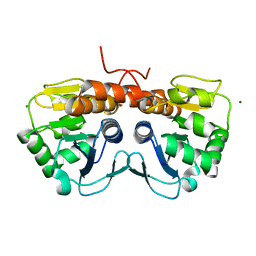 | |
7BCU
 
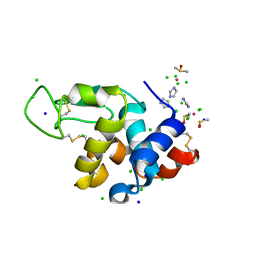 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 1.5 hours. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
8QMJ
 
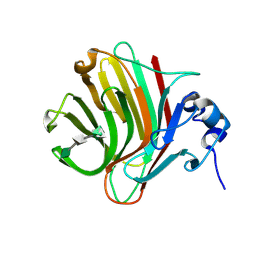 | |
1GHG
 
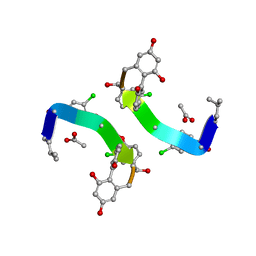 | | CRYSTAL STRUCTURE OF VANCOMYCIN AGLYCON | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, VANCOMYCIN AGLYCON | | Authors: | Kaplan, J, Korty, B.D, Axelsen, P.H, Loll, P.J. | | Deposit date: | 2000-12-13 | | Release date: | 2001-02-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The Role of Sugar Residues in Molecular Recognition by Vancomycin
J.Med.Chem., 44, 2001
|
|
4A7U
 
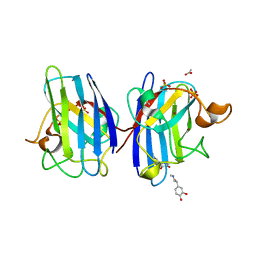 | | Structure of human I113T SOD1 complexed with adrenaline in the p21 space group. | | Descriptor: | ACETATE ION, COPPER (II) ION, L-EPINEPHRINE, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
1V0L
 
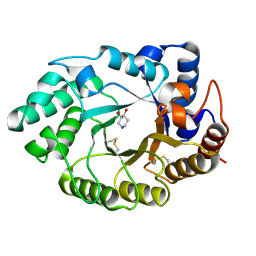 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
3I5E
 
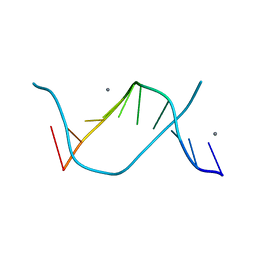 | |
8RBI
 
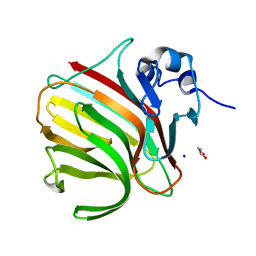 | |
2PNE
 
 | | Crystal Structure of the Snow Flea Antifreeze Protein | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
8RC7
 
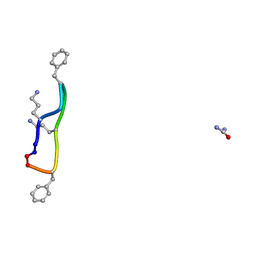 | |
5XJZ
 
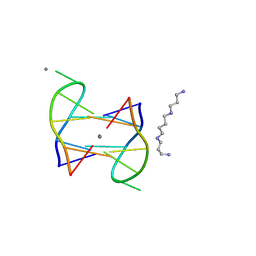 | | Structure of DNA1-Ag complex | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*GP*CP*GP*C)-3'), SILVER ION, SPERMINE | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4F1U
 
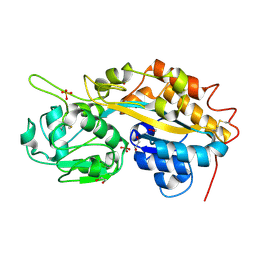 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 4.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, ... | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
3PSM
 
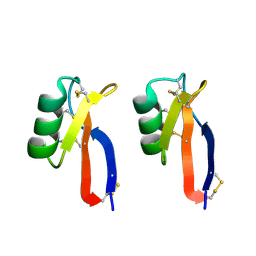 | |
6TN1
 
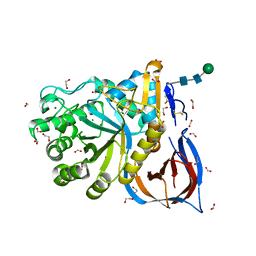 | | Unliganded Crystal Structure of Recombinant GBA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1TQG
 
 | | CheA phosphotransferase domain from Thermotoga maritima | | Descriptor: | Chemotaxis protein cheA | | Authors: | Quezada, C.M, Gradinaru, C, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Helical Shifts Generate Two Distinct Conformers in the Atomic Resolution Structure of the CheA Phosphotransferase Domain from Thermotoga maritima.
J.Mol.Biol., 341, 2004
|
|
4HVU
 
 | |
4HVW
 
 | |
