5XS1
 
 | |
5XZK
 
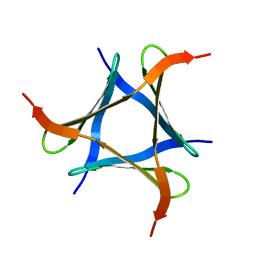 | | Pholiota squarrosa lectin trimer | | Descriptor: | lectin (PhoSL) | | Authors: | Yamasaki, K. | | Deposit date: | 2017-07-12 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The trimeric solution structure and fucose-binding mechanism of the core fucosylation-specific lectin PhoSL.
Sci Rep, 8, 2018
|
|
6UT2
 
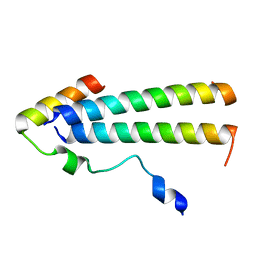 | | 3D structure of the leiomodin/tropomyosin binding interface | | Descriptor: | Leiomodin-2, Tropomyosin alpha-1 chain chimeric peptide | | Authors: | Tolkatchev, D, Smith, G.E, Helms, G.L, Cort, J.R, Kostyukova, A.S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Leiomodin creates a leaky cap at the pointed end of actin-thin filaments.
Plos Biol., 18, 2020
|
|
5UZI
 
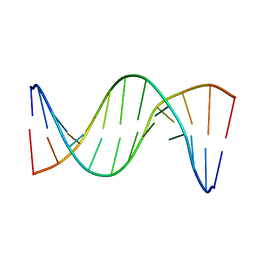 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNAm1A16 structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*(M1A)P*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
6CGH
 
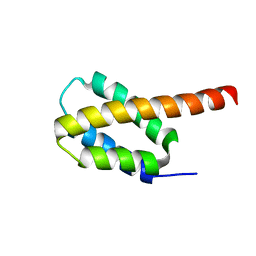 | | Solution structure of the four-helix bundle region of human J-protein Zuotin, a component of ribosome-associated complex (RAC) | | Descriptor: | DnaJ homolog subfamily C member 2 | | Authors: | Shrestha, O.K, Lee, W, Tonelli, M, Cornilescu, G, Markley, J.L, Ciesielski, S.J, Craig, E.A. | | Deposit date: | 2018-02-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and evolution of the 4-helix bundle domain of Zuotin, a J-domain protein co-chaperone of Hsp70.
Plos One, 14, 2019
|
|
5IIV
 
 | | GCN4p pH 1.5 | | Descriptor: | General control protein GCN4 | | Authors: | Brady, M.R, Kaplan, A.R, Alexandrescu, A.T. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structures of GCN4p Are Largely Conserved When Ion Pairs Are Disrupted at Acidic pH but Show a Relaxation of the Coiled Coil Superhelix.
Biochemistry, 56, 2017
|
|
7U8H
 
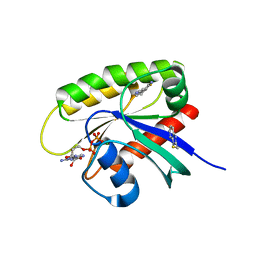 | | Discovery of a KRAS G12V Inhibitor in vivo Tool Compound starting from an HSQC-NMR based Fragment Hit | | Descriptor: | 1H-benzimidazol-2-ylmethanethiol, 2-amino-4,5,6,7-tetrahydro-1-benzothiophene-3-carbonitrile, GTPase KRas, ... | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS G12C Inhibitor.
J.Med.Chem., 65, 2022
|
|
3ZTG
 
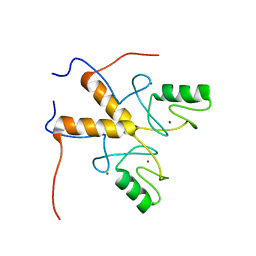 | | Solution structure of the RING finger-like domain of Retinoblastoma Binding Protein-6 (RBBP6) | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE RBBP6, ZINC ION | | Authors: | Kappo, M.A, Ab, E, Atkinson, R.A, Faro, A, Muleya, V, Mulaudzi, T, Poole, J.O, McKenzie, J.M, Pugh, D.J.R. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ring Finger-Like Domain of Retinoblastoma Binding Protein-6 (Rbbp6) Suggests It Functions as a U-Box
J.Biol.Chem., 287, 2012
|
|
4BWH
 
 | |
2YUC
 
 | |
7QJF
 
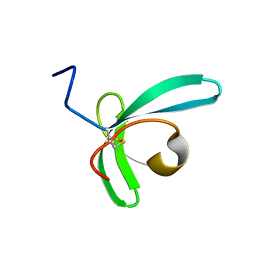 | | Llp mutant C1G, lytic conversion lipoprotein of phage T5 | | Descriptor: | Lytic conversion lipoprotein | | Authors: | Degroux, S, Mestdach, E, Vives, C, Le Roy, A, Salmon, L, Herrman, T, Breyton, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Method: | SOLUTION NMR | | Cite: | Llp mutant C1G, lytic conversion lipoprotein of phage T5
To Be Published
|
|
7QSH
 
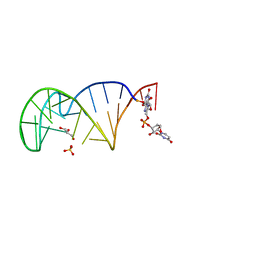 | | 23S ribosomal RNA Sarcin Ricin Loop 27-nt fragment containing a Xanthosine residue at position 2648 | | Descriptor: | 23S ribosomal RNA Sarcin Ricin Loop 27-nucleotide fragment, 9-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-[[tris(oxidanyl)-$l^{5}-phosphanyl]oxymethyl]oxolan-2-yl]-2-oxidanyl-1~{H}-purin-6-one, GLYCEROL, ... | | Authors: | Ennifar, E, Micura, R. | | Deposit date: | 2022-01-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Towards a comprehensive understanding of RNA deamination: synthesis and properties of xanthosine-modified RNA.
Nucleic Acids Res., 50, 2022
|
|
7QUA
 
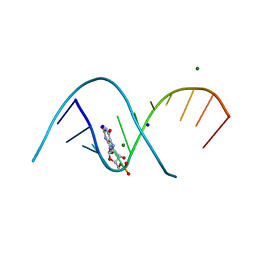 | | Duplex RNA containing Xanthosine-Cytosine base pairs | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*GP*CP*GP*(XAN)P*AP*UP*UP*AP*GP*CP*G)-3'), SODIUM ION | | Authors: | Ennifar, E, Micura, R. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Towards a comprehensive understanding of RNA deamination: synthesis and properties of xanthosine-modified RNA.
Nucleic Acids Res., 50, 2022
|
|
7QTN
 
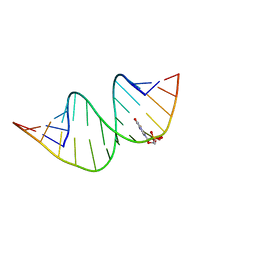 | | Duplex RNA containing Xanthosine-Cytosine base pairs | | Descriptor: | RNA (5'-R(*GP*GP*UP*AP*(RY)P*UP*GP*CP*GP*(XAN)P*UP*AP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*AP*CP*UP*GP*CP*GP*(XAM)P*UP*AP*CP*C)-3') | | Authors: | Ennifar, E, Micura, R. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Towards a comprehensive understanding of RNA deamination: synthesis and properties of xanthosine-modified RNA.
Nucleic Acids Res., 50, 2022
|
|
7QS6
 
 | |
2YUB
 
 | |
8TZM
 
 | |
2YW5
 
 | | Solution structure of the bromodomain from human bromodomain containing protein 3 | | Descriptor: | Bromodomain-containing protein 3 | | Authors: | Furue, M, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-19 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the bromodomain from human bromodomain containing protein 3
To be Published
|
|
6CZT
 
 | |
6D10
 
 | |
6FNV
 
 | |
6K4V
 
 | | The solution structure of the smart chimeric peptide G6 | | Descriptor: | smart chimeric peptide G6 | | Authors: | Wang, J.H, Liu, X.H. | | Deposit date: | 2019-05-27 | | Release date: | 2019-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli.
Commun Biol, 3, 2020
|
|
6K4W
 
 | | smart chimeric peptide SCP-A6 | | Descriptor: | SCP-A6 | | Authors: | Wang, J.H, Liu, X.H. | | Deposit date: | 2019-05-27 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli.
Commun Biol, 3, 2020
|
|
6L95
 
 | | transmembrane-domain of Bax | | Descriptor: | Apoptosis regulator BAX | | Authors: | Bo, O.Y, Fujiao, L. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | transmembrane-domain of Bax
To Be Published
|
|
8C7B
 
 | |
