5CVO
 
 | | WDR48:USP46~ubiquitin ternary complex | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 46, WD repeat-containing protein 48, ... | | Authors: | Harris, S.F, Yin, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.885 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
3EU1
 
 | | Crystal Structure determination of goat hemoglobin (Capra hircus) at 3 angstrom resolution | | Descriptor: | Hemoglobin subunit alpha-1/2, Hemoglobin subunit beta-A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sathya Moorthy, P, Neelagandan, K, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2008-10-09 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Purification, crystallization and preliminary X-ray diffraction studies on goat (Capra hircus) hemoglobin - a low oxygen affinity species
Protein Pept.Lett., 16, 2009
|
|
2ZML
 
 | | Crystal structure of basic winged bean lectin in complex with Gal-ALPHA 1,4 Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZMK
 
 | | Crystl structure of Basic Winged bean lectin in complex with Gal-alpha-1,4-Gal-Beta-Ethylene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7VPN
 
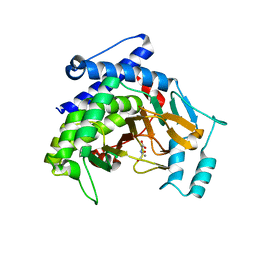 | |
7V7W
 
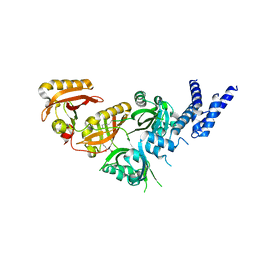 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | Descriptor: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
2AKC
 
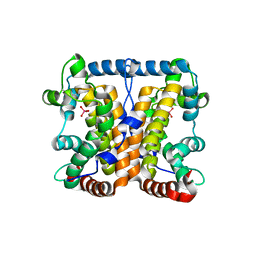 | |
7KBQ
 
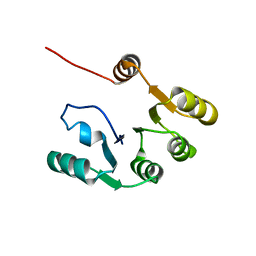 | |
7JZS
 
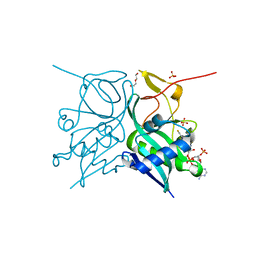 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with CoA | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Park, J, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
7JQ7
 
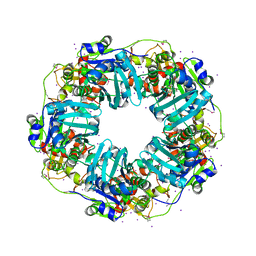 | | The Phi-28 gp11 DNA packaging Motor | | Descriptor: | Encapsidation protein, IODIDE ION, SULFATE ION | | Authors: | Morais, M.C, White, M.A, Dill, E. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Atomistic basis of force generation, translocation, and coordination in a viral genome packaging motor.
Nucleic Acids Res., 49, 2021
|
|
7JQP
 
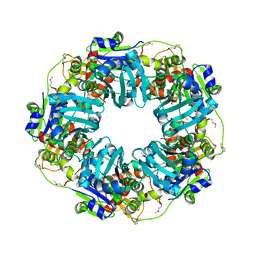 | | The Phi-28 gp11 DNA packaging Motor | | Descriptor: | Encapsidation protein, SULFATE ION | | Authors: | Morais, M.C, White, M.A, Dill, E. | | Deposit date: | 2020-08-11 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Atomistic basis of force generation, translocation, and coordination in a viral genome packaging motor.
Nucleic Acids Res., 49, 2021
|
|
7JQ6
 
 | | The Phi-28 gp11 DNA packaging Motor | | Descriptor: | Encapsidation protein, SULFATE ION | | Authors: | Morais, M.C, White, M.A, Dill, E. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atomistic basis of force generation, translocation, and coordination in a viral genome packaging motor.
Nucleic Acids Res., 49, 2021
|
|
7KD4
 
 | |
1TRB
 
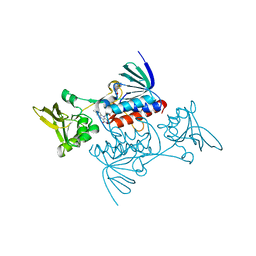 | |
7KD5
 
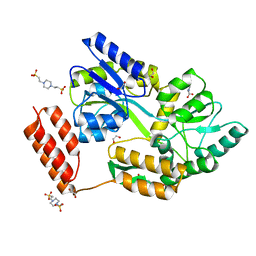 | | Structure of the C-terminal domain of the Menangle virus phosphoprotein (residues 329 -388), fused to MBP. Space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein and Phosphoprotein fusion protein, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Webby, M.N, Kingston, R.L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions.
Viruses, 13, 2021
|
|
1LLA
 
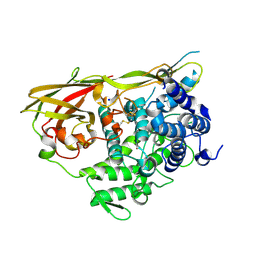 | |
2EFJ
 
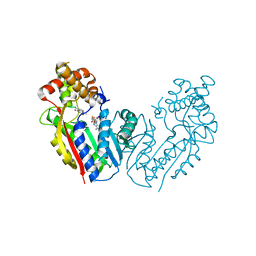 | |
2EG5
 
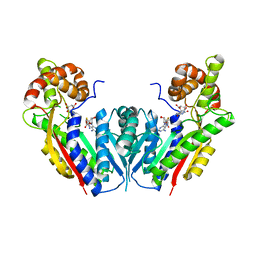 | | The structure of xanthosine methyltransferase | | Descriptor: | 9-[(2R,3R,4S,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)OXOLAN-2-YL]-3H-PURINE-2,6-DIONE, S-ADENOSYL-L-HOMOCYSTEINE, Xanthosine methyltransferase | | Authors: | McCarthy, A.A, McCarthy, J.G. | | Deposit date: | 2007-02-28 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of two N-methyltransferases from the caffeine biosynthetic pathway
Plant Physiol., 144, 2007
|
|
1Q13
 
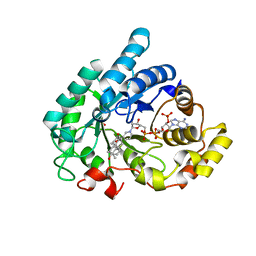 | | Crystal structure of rabbit 20alpha hyroxysteroid dehydrogenase in ternary complex with NADP and testosterone | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin-E2 9-reductase, SULFATE ION, ... | | Authors: | Couture, J.-F, Cantin, L, Legrand, P, Luu-The, V, Labrie, F, Breton, R. | | Deposit date: | 2003-07-18 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Loop relaxation, a mechanism that explains the reduced specificity of rabbit 20alpha-hydroxysteroid dehydrogenase, a member of the aldo-keto reductase superfamily.
J. Mol. Biol., 339, 2004
|
|
2GHP
 
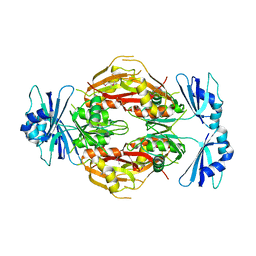 | | Crystal structure of the N-terminal 3 RNA binding domains of the yeast splicing factor Prp24 | | Descriptor: | U4/U6 snRNA-associated splicing factor PRP24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
2E4P
 
 | | Crystal structure of BphA3 (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, SULFATE ION, ... | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2E4Q
 
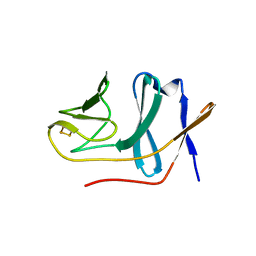 | | Crystal structure of BphA3 (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2ZMN
 
 | | Crystal Structure of basic winged bean lectin in complex with Gal-alpha- 1,6 Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7YSW
 
 | | Cryo-EM Structure of FGF23-FGFR4-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
2YYG
 
 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, SULFATE ION | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
