6SDN
 
 | |
6SDQ
 
 | |
6SFA
 
 | |
6T7N
 
 | |
7DK1
 
 | | Crystal structure of Zinc bound SARS-CoV-2 main protease | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Sonkar, K.S, Panchariya, L, Kuila, S, Khan, W.A, Arockiasamy, A. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Zinc 2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro.
Chem.Commun.(Camb.), 57, 2021
|
|
4B6M
 
 | |
5UMK
 
 | |
5UZY
 
 | |
5V70
 
 | |
5V73
 
 | |
8CZV
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZX
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZW
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8DGY
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d (high resolution) | | Descriptor: | 3C-like proteinase, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{R},2~{R})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-06-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CBR
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBS
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT871 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
7PCL
 
 | | BurG (holo) in complex with 2-hydroxy-2-(hydroxy(isopropyl)amino)acetate (11): Biosynthesis of cyclopropanolrings in bacterial toxins | | Descriptor: | (2S)-2-oxidanyl-2-[oxidanyl(propan-2-yl)amino]ethanoic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
8CZU
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8PUI
 
 | | human PHOX2B C-terminal domain including the polyA fragment at 298K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
8PTL
 
 | | human PHOX2B C-terminal domain including the polyA fragment at 278K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
2Q2R
 
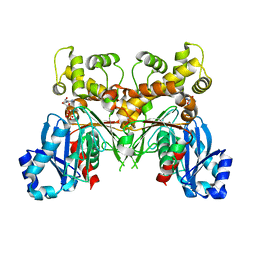 | | Trypanosoma cruzi glucokinase in complex with beta-D-glucose and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucokinase 1, putative, ... | | Authors: | Cordeiro, A.T, Caceres, A.J, Vertommen, D, Concepcion, J.L, Michels, P.A, Versees, W. | | Deposit date: | 2007-05-29 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Trypanosoma cruzi Glucokinase Reveals Features Determining Oligomerization and Anomer Specificity of Hexose-phosphorylating Enzymes.
J.Mol.Biol., 372, 2007
|
|
6O7Y
 
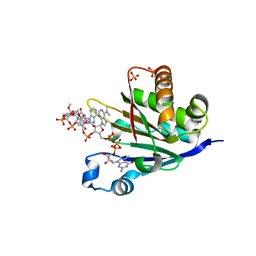 | | Trypanosoma cruzi EIF4E5 translation initiation factor in complex with cap-4 | | Descriptor: | 2-amino-9-[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(S)-({(2R,3R,4R,5R)-3-{[(R)-{[(2R,3R,4R,5R)-3-{[(S)-{[(2R,3R,4R,5R)-5-(4-amino-2-oxopyrimidin-1(2H)-yl)-3-{[(S)-hydroxy{[(2R,3R,4R,5R)-3-hydroxy-4-methoxy-5-(3-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methoxy}phosphoryl]oxy}-4-methoxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-(6-amino-9H-purin-9-yl)-4-methoxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-[6-(dimethylamino)-9H-purin-9-yl]-4-methoxytetrahydrofuran-2-yl}methoxy)(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]-7-methyl-6-oxo-6,9-dihydro-3H-purin-7-ium, Putative Eukaryotic translation initiation factor 4E type 5, SULFATE ION | | Authors: | Guimaraes, B.G, Reolon, L.W. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Trypanosoma cruzi EIF4E5 translation factor homologue in complex with mRNA cap-4.
Nucleic Acids Res., 47, 2019
|
|
6O80
 
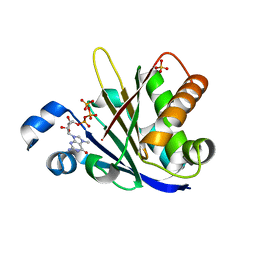 | |
6O7Z
 
 | | Trypanosoma cruzi EIF4E5 translation initiation factor in complex with cap-1 | | Descriptor: | 2-amino-9-[(2R,3R,4S,5R)-5-({[(R)-{[(S)-{[(R)-({(2R,3R,4R,5R)-5-[6-(dimethylamino)-9H-purin-9-yl]-3-hydroxy-4-methoxytetrahydrofuran-2-yl}methoxy)(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]-7-methyl-6-oxo-6,9-dihydro-1H-purin-7-ium, Putative Eukaryotic translation initiation factor 4E type 5, SULFATE ION | | Authors: | Guimaraes, B.G, Reolon, L.W. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Trypanosoma cruzi EIF4E5 translation factor homologue in complex with mRNA cap-4.
Nucleic Acids Res., 47, 2019
|
|
