2KH8
 
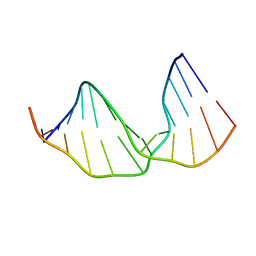 | |
2KHU
 
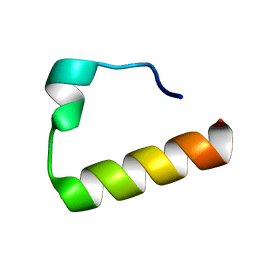 | | Solution Structure of the Ubiquitin-Binding Motif of Human Polymerase Iota | | Descriptor: | Immunoglobulin G-binding protein G, DNA polymerase iota | | Authors: | Bomar, M.G, D'Souza, S, Bienko, M, Dikic, I, Walker, G. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases iota and Rev1.
Mol.Cell, 37, 2010
|
|
2KYY
 
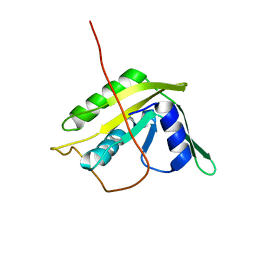 | | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea, Northeast Structural Genomics Consortium Target NeR70A | | Descriptor: | Possible ATP-dependent DNA helicase RecG-related protein | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Maglaqui, M, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea
To be Published
|
|
2L1O
 
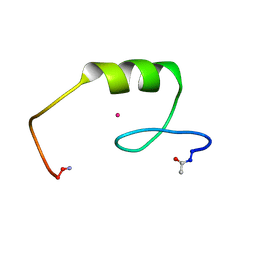 | | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys2His2 zinc finger induces structural rearrangements of typical DNA base determinant positions | | Descriptor: | CADMIUM ION, Transcriptional regulator SUPERMAN | | Authors: | Malgieri, G, Zaccaro, L, Leone, M, Bucci, E, Esposito, S, Baglivo, I, Del Gatto, A, Scandurra, R, Pedone, P.V, Fattorusso, R, Isernia, C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys(2) His(2) zinc finger induces structural rearrangements of typical DNA base determinant positions.
Biopolymers, 95, 2011
|
|
6O3O
 
 | | Structure of human DNAM-1 (CD226) bound to nectin-like protein-5 (necl-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1.
J.Biol.Chem., 294, 2019
|
|
1E0H
 
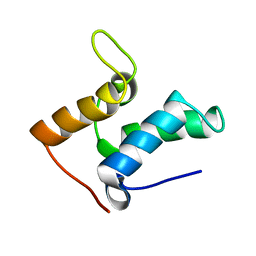 | | Inhibitor Protein Im9 bound to its partner E9 DNase | | Descriptor: | IMMUNITY PROTEIN FOR COLICIN E9 | | Authors: | Boetzel, R, Czisch, M, Kaptein, R, Hemmings, A.M, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 2000-03-28 | | Release date: | 2000-10-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR investigation of the interaction of the inhibitor protein Im9 with its partner DNase.
Protein Sci., 9, 2000
|
|
4RTF
 
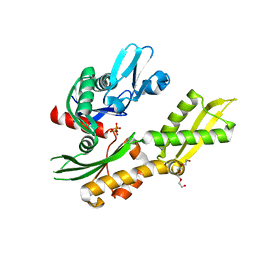 | | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv
To be Published
|
|
7KZU
 
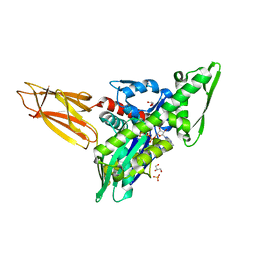 | | Quasi-intermediate state (Q) of a truncated Hsp70 DnaK fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, GLYCEROL, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6Z5N
 
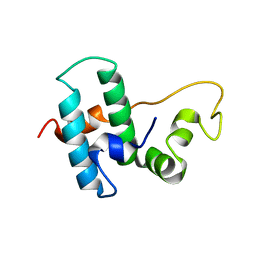 | | DnaJB1 JD-GF | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Avraham-Abayev, M, London, N, Rosenzweig, R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | HSP40 proteins use class-specific regulation to drive HSP70 functional diversity.
Nature, 587, 2020
|
|
7P1E
 
 | | Structure of KDNase from Aspergillus Terrerus in complex with 2,3-difluoro-2-keto-3-deoxynononic acid | | Descriptor: | (2R,3R,4R,5R,6S)-2,3-bis(fluoranyl)-4,5-bis(oxidanyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1B
 
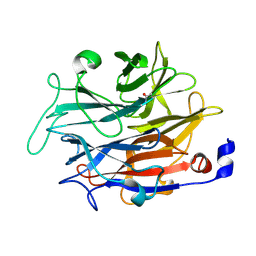 | |
7P1O
 
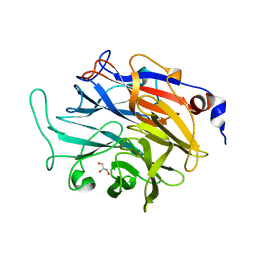 | |
7P1S
 
 | | Structure of KDNase from Trichophyton Rubrum in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, Extracellular sialidase/neuraminidase, SODIUM ION | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1F
 
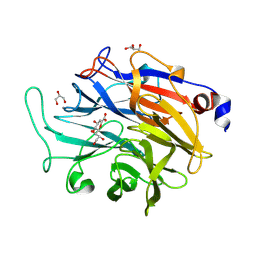 | | Structure of KDNase from Aspergillus terrerus in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1U
 
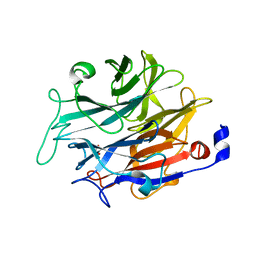 | |
7P1Q
 
 | |
7P1R
 
 | |
7P1D
 
 | |
7P1V
 
 | | Apo structure of KDNase from Trichophyton Rubrum | | Descriptor: | CALCIUM ION, Extracellular sialidase/neuraminidase, GLYCEROL | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
6W6I
 
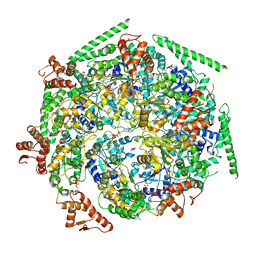 | |
6W6H
 
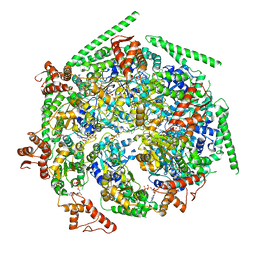 | |
6W6J
 
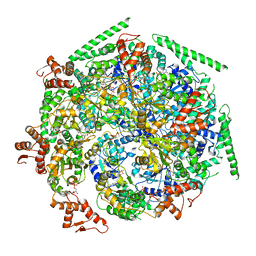 | |
6W6G
 
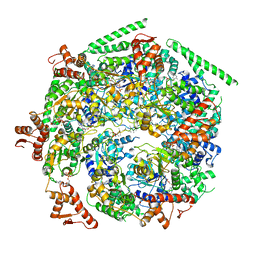 | |
3AT6
 
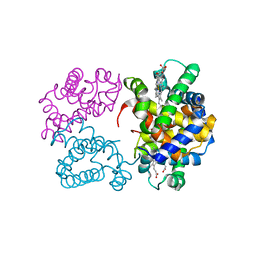 | | Side-necked turtle (Pleurodira, Chelonia, REPTILIA) hemoglobin: cDNA-derived primary structures and X-ray crystal structures of Hb A | | Descriptor: | AlphaA-globin, Beta-globin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasegawa, T, Shishikura, F, Kuwada, T. | | Deposit date: | 2010-12-26 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Side-necked turtle (Pleurodira, Chelonia, reptilia) hemoglobin: cDNA-derived primary structures and X-ray crystal structures of Hb A.
Iubmb Life, 63, 2011
|
|
2XZJ
 
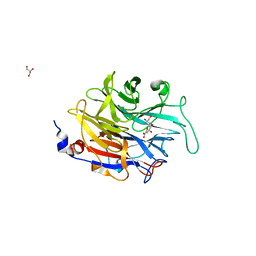 | | THE ASPERGILLUS FUMIGATUS SIALIDASE IS A KDNASE: STRUCTURAL AND MECHANISTIC INSIGHTS | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, EXTRACELLULAR SIALIDASE/NEURAMINIDASE, PUTATIVE, ... | | Authors: | Telford, J.C, Yeung, J.H.F, Kiefel, M.J, Watts, A.G, Hader, S, Chan, J, Bennet, A.J, Moore, M.M, Taylor, G.L. | | Deposit date: | 2010-11-26 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Aspergillus Fumigatus Sialidase is a Kdnase: Structural and Mechanistic Insights.
J.Biol.Chem., 286, 2011
|
|
