7DB0
 
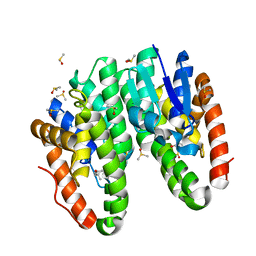 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in dimedone-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAZ
 
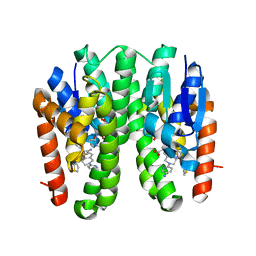 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP015- and GSH-bound form | | Descriptor: | 6-(6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl)pyridin-3-amine, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAY
 
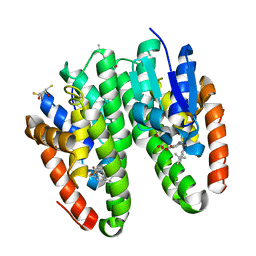 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP013-, and GSH-bound form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-[(5-tert-butyl-2-methyl-phenyl)sulfonylamino]benzoic acid, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB2
 
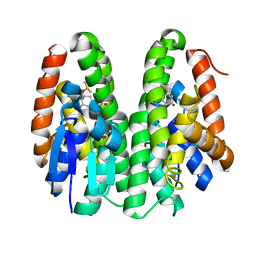 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in GS-dimedone-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB4
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP012- and glutathione-bound form | | Descriptor: | 1-[(4-fluorophenyl)methyl]-2,2-bis(oxidanylidene)thieno[3,2-c][1,2]thiazin-4-one, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAX
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP013-bound form | | Descriptor: | 2-[(5-tert-butyl-2-methyl-phenyl)sulfonylamino]benzoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7000097 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB3
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP011-bound form | | Descriptor: | 4-bromanyl-2-[[2-[(E)-1-(3-methoxyphenyl)ethylideneamino]propan-2-ylamino]methyl]phenol, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
6YGE
 
 | | NADase from Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, AfNADase, ... | | Authors: | Stromland, O, Ziegler, M, Kallio, J.P. | | Deposit date: | 2020-03-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of fungal surface NADases predominantly present in pathogenic species.
Nat Commun, 12, 2021
|
|
7DE8
 
 | |
3Q2L
 
 | | Mouse E-cadherin EC1-2 V81D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, PENTAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
6VR6
 
 | | Structure of ALDH9A1 complexed with NAD+ in space group P1 | | Descriptor: | 4-trimethylaminobutyraldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wyatt, J.W, Tanner, J.J. | | Deposit date: | 2020-02-06 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch.Biochem.Biophys., 691, 2020
|
|
6XWI
 
 | |
8PVM
 
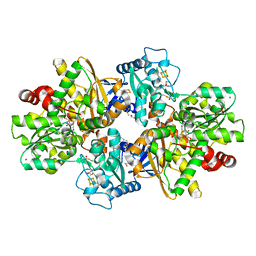 | | formaldehyde-inhibited [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant C299D | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FORMYL GROUP, ... | | Authors: | Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Insights into the Molecular Mechanism of Formaldehyde Inhibition of [FeFe]-Hydrogenases.
J.Am.Chem.Soc., 145, 2023
|
|
1RWZ
 
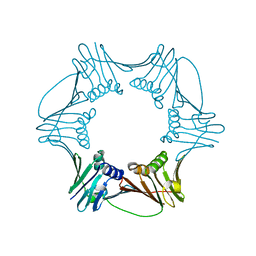 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) from A. fulgidus | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXM
 
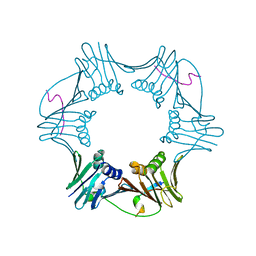 | | C-terminal region of FEN-1 bound to A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, consensus FEN-1 peptide | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
6W5J
 
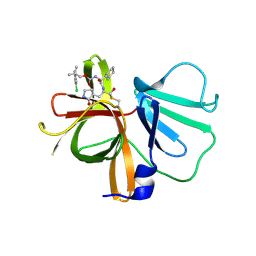 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
3Q2N
 
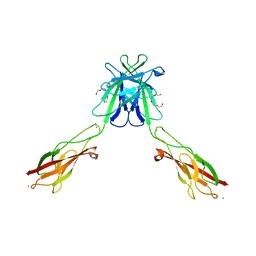 | | Mouse E-cadherin EC1-2 L175D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, TETRAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
8PEC
 
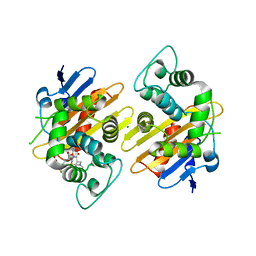 | | OXA-48_Q5-CAZ. Epistasis Arises from Shifting the Rate-Limiting Step during Enzyme Evolution | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION | | Authors: | Leiros, H.-K.S, Frohlich, C. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Epistasis arises from shifting the rate-limiting step during enzyme evolution of a beta-lactamase.
Nat Catal, 7, 2024
|
|
8AJJ
 
 | |
6VTW
 
 | |
8AJK
 
 | | Crystal structure of a C43S variant from the disulfide reductase MerA from Staphylococcus aureus | | Descriptor: | FAD-containing oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Weiland, P, Altegoer, F, Bange, G. | | Deposit date: | 2022-07-28 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MerA functions as a hypothiocyanous acid reductase and defense mechanism in Staphylococcus aureus.
Mol.Microbiol., 119, 2023
|
|
8PEB
 
 | |
8PEA
 
 | |
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
