1NIH
 
 | |
8GPB
 
 | |
5GPB
 
 | |
5ABP
 
 | |
1CA3
 
 | |
6TIM
 
 | |
1FAI
 
 | | THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY | | Descriptor: | IGG2B-KAPPA R19.9 FAB (HEAVY CHAIN), IGG2B-KAPPA R19.9 FAB (LIGHT CHAIN) | | Authors: | Lascombe, M.B, Alzari, P.M, Poljak, R.J, Nisonoff, A. | | Deposit date: | 1992-05-27 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of two crystal forms of FabR19.9 from a monoclonal anti-arsonate antibody.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
9GPB
 
 | |
4GPB
 
 | |
1D81
 
 | |
1GPB
 
 | |
1STP
 
 | |
1LE4
 
 | |
1GPA
 
 | |
6GPB
 
 | |
5TIM
 
 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
2GPB
 
 | |
1LPE
 
 | |
1HSA
 
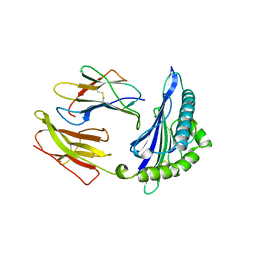 | | THE THREE-DIMENSIONAL STRUCTURE OF HLA-B27 AT 2.1 ANGSTROMS RESOLUTION SUGGESTS A GENERAL MECHANISM FOR TIGHT PEPTIDE BINDING TO MHC | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-B*2705), MODEL PEPTIDE SEQUENCE - ARAAAAAAA | | Authors: | Madden, D.R, Gorga, J.C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1992-08-11 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC.
Cell(Cambridge,Mass.), 70, 1992
|
|
1D82
 
 | |
1HEB
 
 | |
1D58
 
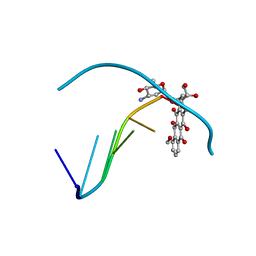 | | THE MOLECULAR STRUCTURE OF A 4'-EPIADRIAMYCIN COMPLEX WITH D(TGATCA) AT 1.7 ANGSTROM RESOLUTION-COMPARISON WITH THE STRUCTURE OF 4'-EPIADRIAMYCIN D(TGTACA) AND D(CGATCG) COMPLEXES | | Descriptor: | 4'-EPIDOXORUBICIN, DNA (5'-D(*TP*GP*AP*TP*CP*A)-3') | | Authors: | Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-02-20 | | Release date: | 1992-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of a 4'-epiadriamycin complex with d(TGATCA) at 1.7A resolution: comparison with the structure of 4'-epiadriamycin d(TGTACA) and d(CGATCG) complexes.
Nucleic Acids Res., 20, 1992
|
|
1HEA
 
 | |
1HEC
 
 | |
1HHP
 
 | |
