1H7J
 
 | | Solution structure of the 26 aa presequence of 5-ALAS | | Descriptor: | AMINOLEVULINIC ACID SYNTHASE 2, ERYTHROID | | Authors: | Goodfellow, B.J, Dias, J.S, Ferreira, G.C, Wray, V, Henklein, P, Macedo, A.L. | | Deposit date: | 2001-07-08 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and Heme Binding of the Presequence of Murine 5-Aminolevulinate Synthase
FEBS Lett., 505, 2001
|
|
1H7K
 
 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Gambarelli, S, Gaillard, J, Sainz, G, Stojanoff, V, Jouve, H.M. | | Deposit date: | 2001-07-08 | | Release date: | 2004-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formation of a Tyrosyl Radical Intermediate in Proteus Mirabilis Catalase by Directed Mutagenesis and Consequences for Nucleotide Reactivity.
Biochemistry, 40, 2001
|
|
1H7L
 
 | | dTDP-MAGNESIUM COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
1H7M
 
 | | Ribosomal Protein L30e from Thermococcus celer | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Chen, Y.W, Wong, K.B. | | Deposit date: | 2001-07-09 | | Release date: | 2003-04-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Ribosomal Protein L30E from the Extreme Thermophile Thermocccus Celer: Thermal Stability and RNA Binding
Biochemistry, 42, 2003
|
|
1H7N
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID AT 1.6 A RESOLUTION | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1H7O
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 5-AMINOLAEVULINIC ACID AT 1.7 A RESOLUTION | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, DELTA-AMINO VALERIC ACID, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1H7P
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 4-KETO-5-AMINO-HEXANOIC (KAH) AT 1.64 A RESOLUTION | | Descriptor: | 5-AMINO-4-HYDROXYHEXANOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1H7Q
 
 | | dTDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, ... | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
1H7R
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH SUCCINYLACETONE AT 2.0 A RESOLUTION. | | Descriptor: | 4,6-DIOXOHEPTANOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1H7S
 
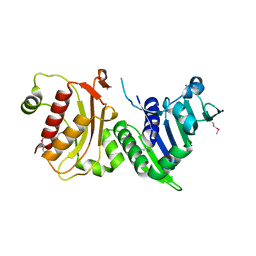 | | N-terminal 40kDa fragment of human PMS2 | | Descriptor: | PMS1 PROTEIN HOMOLOG 2 | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
1H7T
 
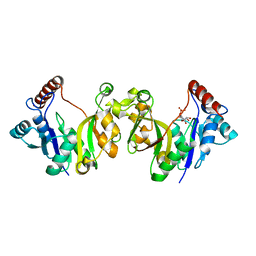 | |
1H7U
 
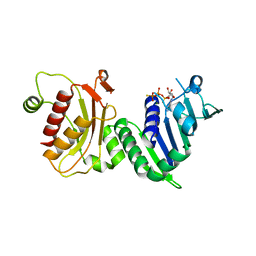 | | hPMS2-ATPgS | | Descriptor: | MAGNESIUM ION, MISMATCH REPAIR ENDONUCLEASE PMS2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
1H7V
 
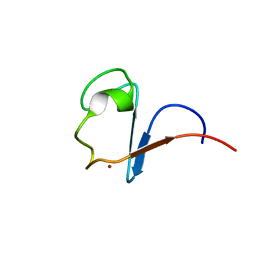 | | Rubredoxin from Guillardia Theta | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Schweimer, K, Hoffmann, S, Wastl, J, Maier, U.G, Roesch, P, Sticht, H. | | Deposit date: | 2001-01-16 | | Release date: | 2002-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a zinc substituted eukaryotic rubredoxin from the cryptomonad alga Guillardia theta.
Protein Sci., 9, 2000
|
|
1H7W
 
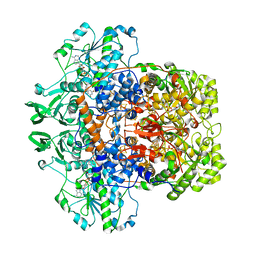 | | Dihydropyrimidine dehydrogenase (DPD) from pig | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
EMBO J., 20, 2001
|
|
1H7X
 
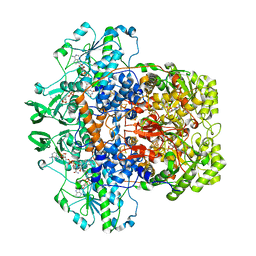 | | Dihydropyrimidine dehydrogenase (DPD) from pig, ternary complex of a mutant enzyme (C671A), NADPH and 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Dihydropyrimidine Dehydrogenase, a Major Determinant of the Pharmacokinetics of the Anti-Cancer Drug 5-Fluorouracil
Embo J., 20, 2001
|
|
1H7Y
 
 | | Translationally Controlled Tumor-associated Protein p23fyp from Schizosaccharomyces pombe | | Descriptor: | TRANSLATIONALLY CONTROLLED TUMOR PROTEIN | | Authors: | Thaw, P, Baxter, N.J, Sedelnikova, S.E, Price, C, Waltho, J.P, Craven, C.J. | | Deposit date: | 2001-01-19 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones.
Nat. Struct. Biol., 8, 2001
|
|
1H7Z
 
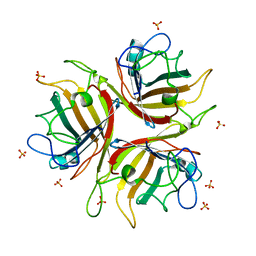 | | Adenovirus Ad3 fibre head | | Descriptor: | ADENOVIRUS FIBRE PROTEIN, SULFATE ION | | Authors: | Durmort, C, Stehlin, C, Schoehn, G, Mitraki, A, Drouet, E, Cusack, S, Burmeister, W.P. | | Deposit date: | 2001-01-21 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Fiber Head of Ad3, a Non-Car-Binding Serotype of Adenovirus
Virology, 285, 2001
|
|
1H80
 
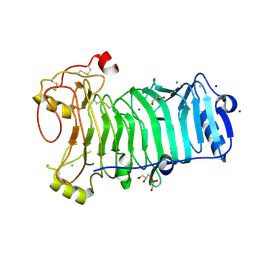 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE- 3,6-ANHYDRO-D-GALACTOSE-2-SULFATE 4 GALACTOHYDROLASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2001-01-22 | | Release date: | 2001-11-27 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Iota-Carrageenase of Alteromonas Fortis. A Beta-Helix Fold-Containing Enzyme for the Degradation of a Highly Polyanionic Polysaccharide
J.Biol.Chem., 276, 2001
|
|
1H81
 
 | | STRUCTURE OF POLYAMINE OXIDASE IN THE REDUCED STATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, POLYAMINE OXIDASE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H82
 
 | | STRUCTURE OF POLYAMINE OXIDASE IN COMPLEX WITH GUAZATINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, N-{8-[(8-{[(E)-AMINO(IMINO)METHYL]AMINO}OCTYL)AMINO]OCTYL}GUANIDINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H83
 
 | | STRUCTURE OF POLYAMINE OXIDASE IN COMPLEX WITH 1,8-DIAMINOOCTANE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, OCTANE 1,8-DIAMINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H84
 
 | | COVALENT ADDUCT BETWEEN POLYAMINE OXIDASE AND N1ethylN11((cycloheptyl)methyl)4,8diazaundecane at pH 4.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-{[3-(METHYLAMINO)PROPYL]AMINO}PROPYL)AMINO]PROPANE-1,1-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H85
 
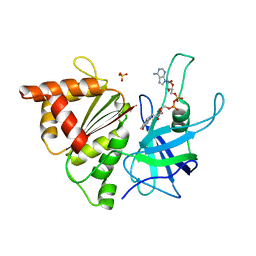 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH VAL 136 REPLACED BY LEU (V136L) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a Cluster of Hydrophobic Residues Near the Fad Cofactor in Anabaena Pcc 7119 Ferredoxin-Nadp+ Reductase for Optimal Complex Formation and Electron Transfer to Ferredoxin
J.Biol.Chem., 276, 2001
|
|
1H86
 
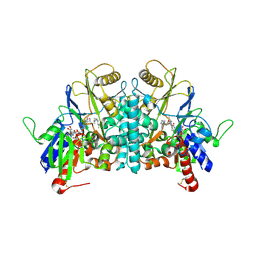 | | COVALENT ADDUCT BETWEEN POLYAMINE OXIDASE AND N1ethylN11((cycloheptyl)methyl)4,8diazaundecane at pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-{[3-(METHYLAMINO)PROPYL]AMINO}PROPYL)AMINO]PROPANE-1,1-DIOL, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H87
 
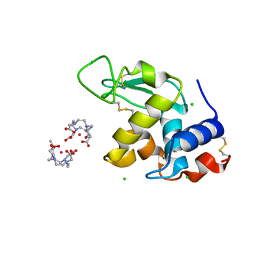 | | Gadolinium derivative of tetragonal Hen Egg-White Lysozyme at 1.7 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Girard, E, Chantalat, L, Vicat, J, Kahn, R. | | Deposit date: | 2001-01-25 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Gd-Hp-Do3A, a Complex to Obtain High-Phasing-Power Heavy Atom Derivatives for Sad and MAD Experiments. Results with Tetragonal Hen Egg-White Lysozyme
Acta Crystallogr.,Sect.D, 58, 2001
|
|
