4AWS
 
 | |
4AWT
 
 | |
4AWU
 
 | |
4AWX
 
 | | Moonlighting functions of FeoC in the regulation of ferrous iron transport in Feo | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, FERROUS IRON TRANSPORT PROTEIN C, NICKEL (II) ION, ... | | Authors: | Hung, K.-W, Tsai, J.-Y, Juan, T.-H, Hsu, Y.-L, Hsiao, C.D, Huang, T.-H. | | Deposit date: | 2012-06-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Klebsiella Pneumoniae Nfeob/Feoc Complex and Roles of Feoc in Regulation of Fe2+ Transport by the Bacterial Feo System.
J.Bacteriol., 194, 2012
|
|
4AWY
 
 | | Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, METALLO-BETA-LACTAMASE AIM-1, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4AWZ
 
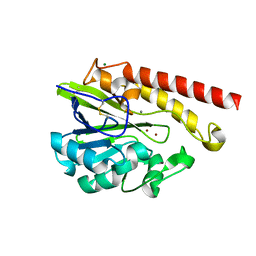 | | Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, METALLO-BETA-LACTAMASE AIM-1, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157.
Antimicrob.Agents Chemother., 56, 2012
|
|
4AX0
 
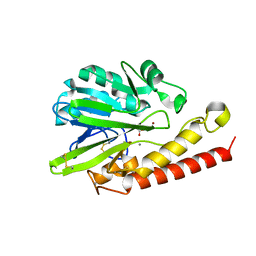 | | Q157A mutant. Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | ACETATE ION, CALCIUM ION, METALLO-BETA-LACTAMASE AIM-1, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4AX1
 
 | | Q157N mutant. Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | ACETATE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4AX2
 
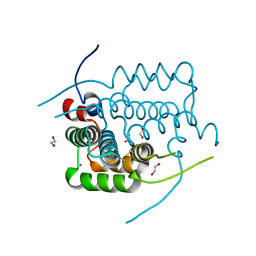 | | New Type VI-secreted toxins and self-resistance proteins in Serratia marcescens | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, RAP1B | | Authors: | English, G, Trunk, K, Rao, V.A, Srikannathasan, V, Fritsch, M.J, Guo, M, Hunter, W.N, Coulthurst, S.J. | | Deposit date: | 2012-06-07 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | New Secreted Toxins and Immunity Proteins Encoded within the Type Vi Secretion System Gene Cluster of Serratia Marcescens
Mol.Microbiol., 86, 2012
|
|
4AX3
 
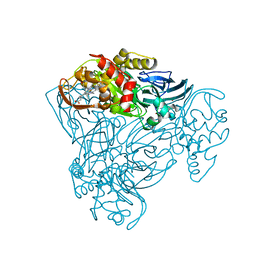 | | Structure of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.6 A resolution | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Cong, H, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2012-06-07 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
4AX4
 
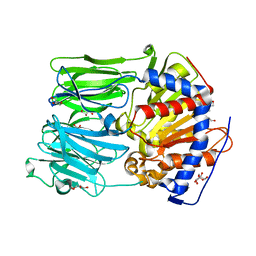 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, H680A MUTANT | | Descriptor: | GLYCEROL, PROLYL OLIGOPEPTIDASE | | Authors: | Szeltner, Z, Juhasz, T, Szamosi, I, Rea, D, Fulop, V, Modos, K, Juliano, L, Polgar, L. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Loops Facing the Active Site of Prolyl Oligopeptidase are Crucial Components in Substrate Gating and Specificity.
Biochim.Biophys.Acta, 1834, 2012
|
|
4AX6
 
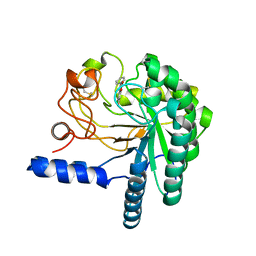 | | HYPOCREA JECORINA CEL6A D221A MUTANT SOAKED WITH 6-CHLORO-4- PHENYLUMBELLIFERYL-BETA-CELLOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloranyl-7-oxidanyl-4-phenyl-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-10 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AX7
 
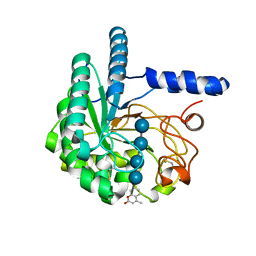 | | Hypocrea jecorina Cel6A D221A mutant soaked with 4-Methylumbelliferyl- beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-11 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AX8
 
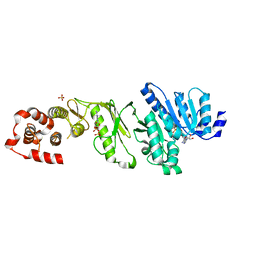 | | Medium resolution structure of the bifunctional kinase- methyltransferase WbdD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization, Dehydration and Experimental Phasing of Wbdd, a Bifunctional Kinase and Methyltransferase from Escherichia Coli O9A.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AX9
 
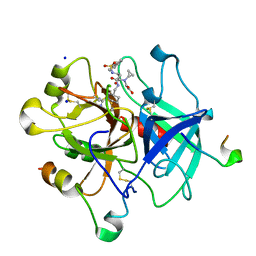 | | Human thrombin complexed with Napsagatran, RO0466240 | | Descriptor: | 2-[[(2S)-4-[[(3S)-1-carbamimidoylpiperidin-3-yl]methylamino]-2-(naphthalen-2-ylsulfonylamino)-4-oxidanylidene-butanoyl] -cyclopropyl-amino]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, ... | | Authors: | Banner, D.W, D'Arcy, A, Winkler, F.K, Hilpert, K, Spinelli, S, Cambillau, C. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Potent and Highly Selective Thrombin Inhibitors.
J.Med.Chem., 37, 1994
|
|
4AXA
 
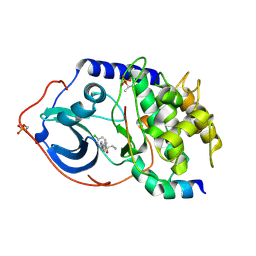 | | Structure of PKA-PKB chimera complexed with (1S)-2-amino-1-(4- chlorophenyl)-1-(4-(1H-pyrazol-4-yl)phenyl)ethan-1-ol | | Descriptor: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Yap, T.A, Walton, M.I, Grimshaw, K.M, tePoele, R.H, Eve, P.D, Valenti, M.R, deHavenBrandon, A.K, Martins, V, Zetterlund, A, Heaton, S.P, Heinzmann, K, Jones, P.S, Feltell, R.E, Reule, M, Woodhead, S.J, Lyons, J.F, Raynaud, F.I, Eccles, S.A, Workman, P, Thompson, N.T, Garrett, M.D. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | At13148 is a Novel, Oral Multi-Agc Kinase Inhibitor with Potent Pharmacodynamic and Antitumor Activity.
Clin.Cancer Res., 18, 2012
|
|
4AXB
 
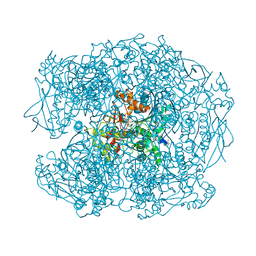 | | Crystal structure of soman-aged human butyrylcholinesterase in complex with 2-PAM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wandhammer, M, de Koning, M, Noort, D, Goeldner, M, Nachon, F. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Step Toward the Reactivation of Aged Cholinesterases -Crystal Structure of Ligands Binding to Aged Human Butyrylcholinesterase
Chem.Biol.Interact, 203, 2013
|
|
4AXC
 
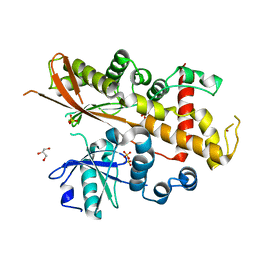 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase apo form | | Descriptor: | GLYCEROL, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, SULFATE ION, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXD
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with AMPPNP | | Descriptor: | CITRIC ACID, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXE
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, SULFATE ION, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXF
 
 | | InsP5 2-K in complex with Ins(3,4,5,6)P4 plus AMPPNP | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, Myo inositol 3,4,5,6 tetrakisphosphate, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXG
 
 | | Structure of eIF4E-Cup complex | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, PROTEIN CUP | | Authors: | Kinkelin, K, Veith, K, Gruenwald, M, Bono, F. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Minimal Eif4E-Cup Complex Revelas a General Mechanism of Eif4E Regulation in Translational Repression
RNA, 18, 2012
|
|
4AXH
 
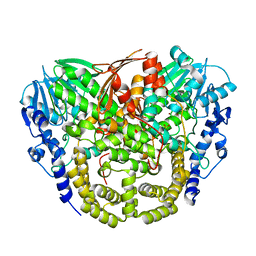 | | Structure and mechanism of the first inverting alkylsulfatase specific for secondary alkylsulfatases | | Descriptor: | SEC-ALKYLSULFATASE, SULFATE ION, ZINC ION | | Authors: | Knaus, T, Schober, M, Faber, K, Macheroux, P, Wagner, U.G. | | Deposit date: | 2012-06-13 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Mechanism of an Inverting Alkylsulfatase from Pseudomonas Sp. Dsm6611 Specific for Secondary Alkylsulfates.
FEBS J., 279, 2012
|
|
4AXI
 
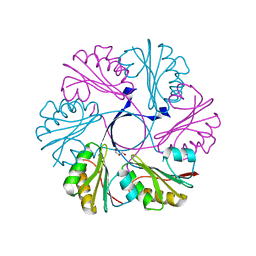 | | Structure of the Clostridium difficile EutS protein | | Descriptor: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, GLYCEROL | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4AXJ
 
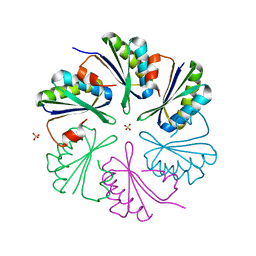 | | Structure of the Clostridium difficile EutM protein | | Descriptor: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, SULFATE ION | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
