1BEU
 
 | | TRP SYNTHASE (D60N-IPP-SER) WITH K+ | | Descriptor: | INDOLE-3-PROPANOL PHOSPHATE, POTASSIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Mozzarelli, A, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryocrystallography and microspectrophotometry of a mutant (alpha D60N) tryptophan synthase alpha 2 beta 2 complex reveals allosteric roles of alpha Asp60.
Biochemistry, 37, 1998
|
|
1BE0
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING ACETIC ACID | | Descriptor: | ACETATE ION, ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1BEX
 
 | | STRUCTURE OF RUTHENIUM-MODIFIED PSEUDOMONAS AERUGINOSA AZURIN | | Descriptor: | AZURIN, COPPER (II) ION, RUTHEMIUM BIS(2,2'-BIPYRIDINE)-2-IMIDAZOLE | | Authors: | Faham, S, Day, M.W, Rees, D.C. | | Deposit date: | 1998-05-18 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ruthenium-modified Pseudomonas aeruginosa azurin and [Ru(2,2'-bipyridine)2(imidazole)2]SO4 x 10H2O.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1KI6
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH A 5-IODOURACIL ANHYDROHEXITOL NUCLEOSIDE | | Descriptor: | 1',5'-ANHYDRO-2',3'-DIDEOXY-2'-(5-IODOURACIL-1-YL)-D-ABABINO-HEXITOL, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Herdewijn, P, Ostrowski, T, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
16PK
 
 | | PHOSPHOGLYCERATE KINASE FROM TRYPANOSOMA BRUCEI BISUBSTRATE ANALOG | | Descriptor: | 1,1,5,5-TETRAFLUOROPHOSPHOPENTYLPHOSPHONIC ACID ADENYLATE ESTER, 3-PHOSPHOGLYCERATE KINASE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | | Authors: | Bernstein, B.E, Bressi, J, Blackburn, M, Gelb, M, Hol, W.G.J. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bisubstrate analog induces unexpected conformational changes in phosphoglycerate kinase from Trypanosoma brucei.
J.Mol.Biol., 279, 1998
|
|
1KI4
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOTHIENYLDEOXYURIDINE | | Descriptor: | 5-BROMOTHIENYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1BEZ
 
 | | HALOALKANE DEHALOGENASE MUTANT WITH TRP 175 REPLACED BY TYR AT PH 5 | | Descriptor: | ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1BE5
 
 | | STRUCTURAL STUDIES OF A STABLE PARALLEL-STRANDED DNA DUPLEX INCORPORATING ISOGUANINE:CYTOSINE AND ISOCYTOSINE:GUANINE BASE PAIRS BY NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA DUPLEX (TGCACGGACT) | | Authors: | Yang, X.-L, Sugiyama, H, Ikeda, S, Saito, I, Wang, A.H.-J. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a stable parallel-stranded DNA duplex incorporating isoguanine:cytosine and isocytosine:guanine basepairs by nuclear magnetic resonance spectroscopy.
Biophys.J., 75, 1998
|
|
1BE3
 
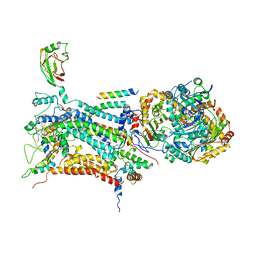 | | CYTOCHROME BC1 COMPLEX FROM BOVINE | | Descriptor: | CYTOCHROME BC1 COMPLEX, FE2/S2 (INORGANIC) CLUSTER, HEME C, ... | | Authors: | Iwata, S, Lee, J.W, Okada, K, Lee, J.K, Iwata, M, Ramaswamy, S, Jap, B.K. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex.
Science, 281, 1998
|
|
1BE4
 
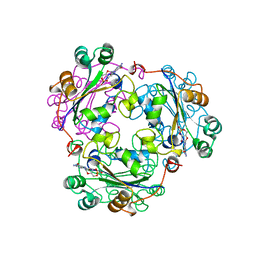 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM B FROM BOVINE RETINA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleoside diphosphate kinase from bovine retina: purification, subcellular localization, molecular cloning, and three-dimensional structure.
Biochemistry, 37, 1998
|
|
1BE1
 
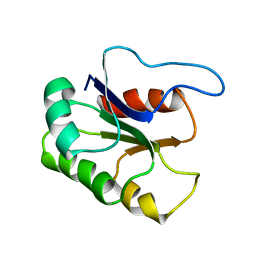 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Tollinger, M, Konrat, R, Hilbert, B.H, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum
Structure, 6, 1998
|
|
1C3D
 
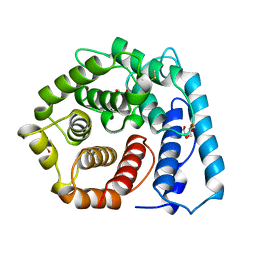 | | X-RAY CRYSTAL STRUCTURE OF C3D: A C3 FRAGMENT AND LIGAND FOR COMPLEMENT RECEPTOR 2 | | Descriptor: | C3D, GLYCEROL | | Authors: | Nagar, B, Jones, R.G, Diefenbach, R.J, Isenman, D.E, Rini, J.M. | | Deposit date: | 1998-05-19 | | Release date: | 1998-10-07 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2.
Science, 280, 1998
|
|
2GPR
 
 | | GLUCOSE PERMEASE IIA FROM MYCOPLASMA CAPRICOLUM | | Descriptor: | GLUCOSE-PERMEASE IIA COMPONENT | | Authors: | Huang, K, Herzberg, O. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A promiscuous binding surface: crystal structure of the IIA domain of the glucose-specific permease from Mycoplasma capricolum.
Structure, 6, 1998
|
|
1BE2
 
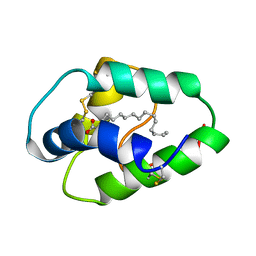 | | LIPID TRANSFER PROTEIN COMPLEXED WITH PALMITATE, NMR, 10 STRUCTURES | | Descriptor: | LIPID TRANSFER PROTEIN, PALMITIC ACID | | Authors: | Lerche, M.H, Poulsen, F.M. | | Deposit date: | 1998-05-19 | | Release date: | 1998-12-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of barley lipid transfer protein complexed with palmitate. Two different binding modes of palmitate in the homologous maize and barley nonspecific lipid transfer proteins.
Protein Sci., 7, 1998
|
|
1FZC
 
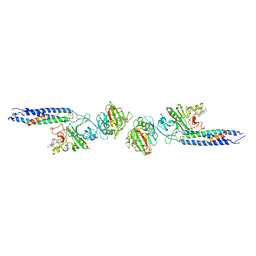 | | CRYSTAL STRUCTURE OF FRAGMENT DOUBLE-D FROM HUMAN FIBRIN WITH TWO DIFFERENT BOUND LIGANDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Everse, S.J, Spraggon, G, Veerapandian, L, Riley, M, Doolittle, R.F. | | Deposit date: | 1998-05-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of fragment double-D from human fibrin with two different bound ligands.
Biochemistry, 37, 1998
|
|
1SKP
 
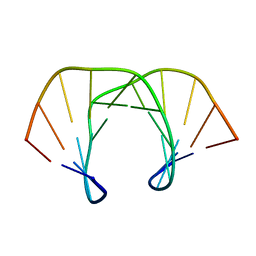 | | NMR STRUCTURE OF D(GCATATGATAG)(DOT)D(CTATCATATGC): A CONSENSUS SEQUENCE FOR PROMOTERS RECOGNIZED BY SIGMA-K RNA POLYMERASE, 4 STRUCTURES | | Descriptor: | SIGMA-K RNA POLYMERASE CONSENSUS SEQUENCE | | Authors: | Tonelli, M, Ragg, E, Bianucci, A.M, Lesiak, K, James, T.L. | | Deposit date: | 1998-05-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of d(GCATATGATAG). d(CTATCATATGC): a consensus sequence for promoters recognized by sigma K RNA polymerase.
Biochemistry, 37, 1998
|
|
7MSF
 
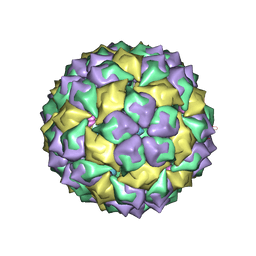 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*AP*AP*CP*AP*GP*GP*CP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-20 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
1BE8
 
 | | TRANS-CINNAMOYL-SUBTILISIN IN WATER | | Descriptor: | CALCIUM ION, PHENYLETHYLENECARBOXYLIC ACID, SUBTILISIN CARLSBERG | | Authors: | Schmitke, J.L, Stern, L.J, Klibanov, A.M. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of x-ray crystal structures of an acyl-enzyme intermediate of subtilisin Carlsberg formed in anhydrous acetonitrile and in water.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BE9
 
 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BE7
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
1BE6
 
 | | TRANS-CINNAMOYL-SUBTILISIN IN ANHYDROUS ACETONITRILE | | Descriptor: | ACETONITRILE, CALCIUM ION, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Schmitke, J.L, Stern, L.J, Klibanov, A.M. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of x-ray crystal structures of an acyl-enzyme intermediate of subtilisin Carlsberg formed in anhydrous acetonitrile and in water.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BFO
 
 | |
1BFE
 
 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
2EZP
 
 | |
2EZS
 
 | |
