4LDQ
 
 | |
4E2F
 
 | |
1KYZ
 
 | | Crystal Structure Analysis of Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase Ferulic Acid Complex | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
2WQV
 
 | |
2HD9
 
 | | Crystal structure of PH1033 from Pyrococcus horikoshii OT3 | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Sugahara, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-20 | | Release date: | 2006-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nucleant-mediated protein crystallization with the application of microporous synthetic zeolites.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2VOC
 
 | | THIOREDOXIN A ACTIVE SITE MUTANTS FORM MIXED DISULFIDE DIMERS THAT RESEMBLE ENZYME-SUBSTRATE REACTION INTERMEDIATE | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOREDOXIN | | Authors: | Kouwen, T.R.H.M, Andrell, J, Schrijver, R, Dubois, J.Y.F, Maher, M.J, Iwata, S, Carpenter, E.P, van Dijl, J.M. | | Deposit date: | 2008-02-13 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thioredoxin A active-site mutants form mixed disulfide dimers that resemble enzyme-substrate reaction intermediates.
J. Mol. Biol., 379, 2008
|
|
2WQU
 
 | |
3AQ0
 
 | | Ligand-bound form of Arabidopsis medium/long-chain length prenyl pyrophosphate synthase (surface polar residue mutant) | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, DI(HYDROXYETHYL)ETHER, FARNESYL DIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase
Plant Physiol., 155, 2011
|
|
4RMK
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in P65 crystal form | | Descriptor: | CALCIUM ION, Latrophilin-3 | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
3RDD
 
 | | Human Cyclophilin A Complexed with an Inhibitor | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-04-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
1QXT
 
 | | Crystal structure of precyclized intermediate for the green fluorescent protein R96A variant (A) | | Descriptor: | green-fluorescent protein | | Authors: | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-09-08 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures
Proc.Natl.Acad.Sci.USA, 100
|
|
3CMC
 
 | | Thioacylenzyme intermediate of Bacillus stearothermophilus phosphorylating GAPDH | | Descriptor: | 1,2-ETHANEDIOL, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Moniot, S, Vonrhein, C, Bricogne, G, Didierjean, C, Corbier, C. | | Deposit date: | 2008-03-21 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Trapping of the Thioacylglyceraldehyde-3-phosphate Dehydrogenase Intermediate from Bacillus stearothermophilus: DIRECT EVIDENCE FOR A FLIP-FLOP MECHANISM
J.Biol.Chem., 283, 2008
|
|
4JE9
 
 | | Crystal structure of an engineered metal-free RIDC1 construct with four interfacial disulfide bonds | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562 | | Authors: | Tezcan, F.A, Medina-Morales, A.M, Perez, A, Brodin, J.D. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | In Vitro and Cellular Self-Assembly of a Zn-Binding Protein Cryptand via Templated Disulfide Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
3TS1
 
 | |
2ZBN
 
 | |
2TCT
 
 | |
3AKB
 
 | | Structural basis for prokaryotic calcium-mediated regulation by a Streptomyces coelicolor calcium-binding protein | | Descriptor: | CALCIUM ION, Putative calcium binding protein | | Authors: | Zhao, X, Pang, H, Wang, S, Zhou, W, Yang, K, Bartlam, M. | | Deposit date: | 2010-07-09 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for prokaryotic calciummediated regulation by a Streptomyces coelicolor calcium binding protein
Protein Cell, 1, 2010
|
|
3O31
 
 | | E81Q mutant of MtNAS in complex with a reaction intermediate | | Descriptor: | BROMIDE ION, N-[(3S)-3-amino-3-carboxypropyl]-L-glutamic acid, ThermoNicotianamine Synthase | | Authors: | Dreyfus, C, Pignol, D, Arnoux, P. | | Deposit date: | 2010-07-23 | | Release date: | 2011-06-08 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystallographic structure of thermoNicotianamine synthase with a synthetic reaction intermediate highlights the sequential processing mechanism.
Chem.Commun.(Camb.), 47, 2011
|
|
8R2I
 
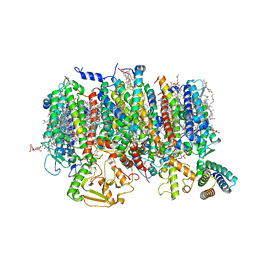 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | Deposit date: | 2023-11-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
2YCE
 
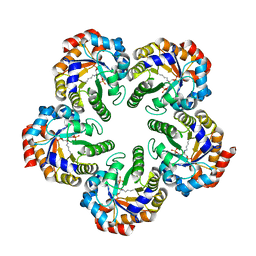 | | Structure of an Archaeal fructose-1,6-bisphosphate aldolase with the catalytic Lys covalently bound to the carbinolamine intermediate of the substrate. | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS 1 | | Authors: | Lorentzen, E, Siebers, B, Hensel, R, Pohl, E. | | Deposit date: | 2011-03-14 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of the Schiff Base Forming Fructose-1,6-Bisphosphate Aldolase: Structural Analysis of Reaction Intermediates.
Biochemistry, 44, 2005
|
|
4BOF
 
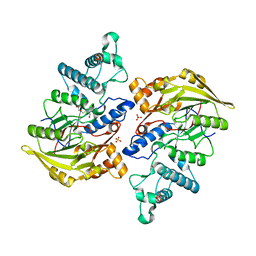 | | Crystal structure of arginine deiminase from group A streptococcus | | Descriptor: | ARGININE DEIMINASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Henningham, A, Ericsson, D.J, Langer, K, Casey, L, Jovcevski, B, Chhatwal, G.S, Aquilina, J.A, Batzloff, M.R, Kobe, B, Walker, M.J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-informed design of an enzymatically inactive vaccine component for group A Streptococcus.
MBio, 4, 2013
|
|
2PMK
 
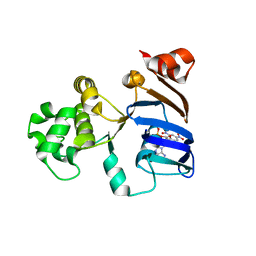 | | Crystal structures of an isolated ABC-ATPase in complex with TNP-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE | | Authors: | Oswald, C, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2007-04-23 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Water-mediated protein-fluorophore interactions modulate the affinity of an ABC-ATPase/TNP-ADP complex
J.Struct.Biol., 162, 2008
|
|
1UMD
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
6S8C
 
 | |
