4PN8
 
 | | A de novo designed pentameric coiled coil CC-Pent. | | Descriptor: | CC-Pent | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
8ATV
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
1OBG
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHORIBOSYLAMIDOIMIDAZOLE- SUCCINOCARBOXAMIDE SYNTHASE, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
1OBD
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
4PN9
 
 | | A de novo designed hexameric coiled coil CC-Hex2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CC-Hex2 | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
4PNA
 
 | | A de novo designed heptameric coiled coil CC-Hept | | Descriptor: | CC-Hept, GLYCEROL | | Authors: | Burton, A.J, Wood, C.W, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
4PND
 
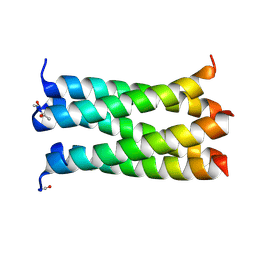 | | A de novo designed pentameric coiled coil CC-Pent_Variant | | Descriptor: | CC-Pent_Variant | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
8C5U
 
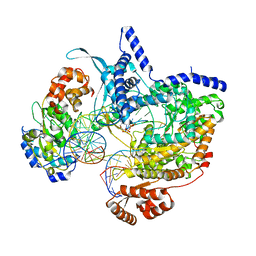 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG (IC8) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8C5S
 
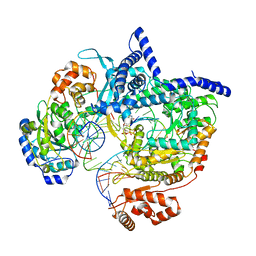 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 7-mer RNA, pppGpGpUpApApApU (IC7) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
4PNB
 
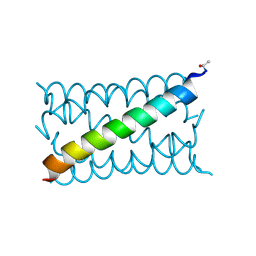 | | A de novo designed hexameric coiled coil CC-Hex3. | | Descriptor: | CC-Hex3 | | Authors: | Wood, C.W, Burton, A.J, Thomson, A.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-22 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Computational design of water-soluble alpha-helical barrels.
Science, 346, 2014
|
|
4Q4Z
 
 | | Thermus thermophilus RNA polymerase de novo transcription initiation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ADENOSINE-5'-TRIPHOSPHATE, DNA (25-MER), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | SULFATE ION, a novel designed pore protein, affinity purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
1CHU
 
 | |
6TVJ
 
 | |
3R2X
 
 | |
1DB3
 
 | | E.COLI GDP-MANNOSE 4,6-DEHYDRATASE | | Descriptor: | GDP-MANNOSE 4,6-DEHYDRATASE | | Authors: | Somoza, J.R, Menon, S, Somers, W.S, Sullivan, F.X. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of Escherichia coli GDP-mannose 4,6 dehydratase provides insights into the enzyme's catalytic mechanism and regulation by GDP-fucose.
Structure Fold.Des., 8, 2000
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5IEN
 
 | | Structure of CDL2.2, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.2, GLYCEROL | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IEO
 
 | | Structure of CDL2.3a, a computationally designed Vitamin-D3 binder | | Descriptor: | 1,2-ETHANEDIOL, 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3a | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IEP
 
 | | Structure of CDL2.3b, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3b | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
7BAY
 
 | |
7DCL
 
 | |
4XTD
 
 | | Structure of the covalent intermediate E-XMP* of the IMP dehydrogenase of Ashbya gossypii | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-01-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
