5A03
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
6S5M
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-n-propyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-propyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
8Y1Z
 
 | | Crystal structure of the Mcl-1 in complex with a Short BH3 peptide of BAK | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Short BH3 peptide from Bcl-2 homologous antagonist/killer | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Deciphering molecular specificity in MCL-1/BAK interaction and its implications for designing potent MCL-1 inhibitors.
Cell Death Differ., 32, 2025
|
|
9HBQ
 
 | |
9HDN
 
 | |
9HD8
 
 | | SARS-CoV-2 Main Protease in complex with with (1R)-N-(3-chlorophenyl)-N-[4-(2,4-dioxo-1H-pyrimidin-5-yl)phenyl]-3-oxo-indane-1-carboxamide | | Descriptor: | (1~{R})-~{N}-[4-[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-5-yl]phenyl]-~{N}-(3-chlorophenyl)-3-oxidanylidene-1,2-dihydroindene-1-carboxamide, 3C-like proteinase nsp5, SODIUM ION | | Authors: | Hanoulle, X, Charton, J, Deprez, B. | | Deposit date: | 2024-11-12 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | SARS-CoV-2 Main Protease in complex with with (1R)-N-(3-chlorophenyl)-N-[4-(2,4-dioxo-1H-pyrimidin-5-yl)phenyl]-3-oxo-indane-1-carboxamide
To Be Published
|
|
8Y1Y
 
 | | Crystal structure of the Mcl-1 in complex with a long BH3 peptide of BAK | | Descriptor: | BH3 peptide from Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Deciphering molecular specificity in MCL-1/BAK interaction and its implications for designing potent MCL-1 inhibitors.
Cell Death Differ., 32, 2025
|
|
8AB2
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
5ODR
 
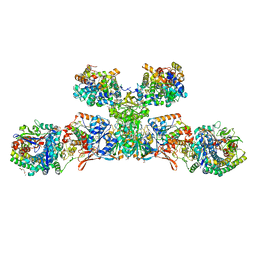 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 2 minutes. | | Descriptor: | 1-THIOETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
9C0U
 
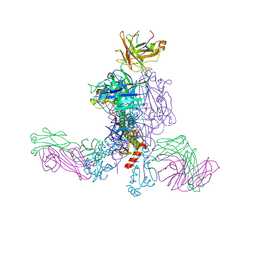 | | Crystal structure of chimeric hemagglutinin cH5/1 in complex with broad protective antibody 31.b.09 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 31.b.09 Fab heavy chain, ... | | Authors: | Nguyen, T.K.Y, Zhu, X, Wilson, I.A. | | Deposit date: | 2024-05-27 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural characterization of influenza group 1 chimeric hemagglutinins as broad vaccine immunogens.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6FWL
 
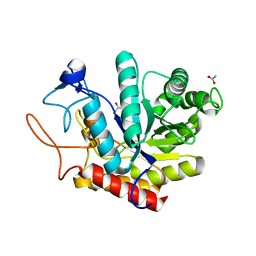 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex alpha-Glc-1,3-(1,2-anhydro-carba-mannose) | | Descriptor: | (1~{S},2~{S},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
9C22
 
 | |
8K73
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((1-(phenylsulfonyl)pyrrolidine-3-carbonyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-3-oxidanyl-2-[(3~{R})-1-(phenylsulfonyl)pyrrolidin-3-yl]carbonylimino-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
5THT
 
 | | Crystal Structure of G303A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
6H6Z
 
 | |
5EUN
 
 | | The Crystal Structure of Human Kynurenine Aminotransferase II, PLP-bound form, at 1.83 A | | Descriptor: | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Nematollahi, A, Sun, G, Kwan, A, Jeffries, C.M, Harrop, S.J, Hanrahan, J.R, Nadvi, N.A, Church, W.B. | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-16 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 angstrom Resolution.
Int J Mol Sci, 17, 2016
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
8K1H
 
 | |
6JCG
 
 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
8K1G
 
 | |
4O06
 
 | | 1.15A Resolution Structure of the Proteasome Assembly Chaperone Nas2 PDZ Domain | | Descriptor: | Probable 26S proteasome regulatory subunit p27, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Singh, C.R, Chowdhury, W.Q, Geanes, E, Roelofs, J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | 1.15 angstrom resolution structure of the proteasome-assembly chaperone Nas2 PDZ domain.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5ZPW
 
 | | Generation of a long-acting fusion inhibitor against HIV-1 | | Descriptor: | MET-THR-TRP-GLU-GLU-TRP-ASP-MK8-LYS-ILE-GLU-MK8-TYR-THR-MK8-LYS-ILE-GLU-MK8-LEU-ILE-LYS-LYS-SER, Transmembrane protein gp41 | | Authors: | Guo, Y, Shi, X.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Generation of a long-acting fusion inhibitor against HIV-1.
Medchemcomm, 9, 2018
|
|
6RKG
 
 | | 1.32 A RESOLUTION OF SPOROSARCINA PASTEURII UREASE INHIBITED IN THE PRESENCE OF NBPTO AT pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Impact of pH on Catalytically Critical Protein Conformational Changes: The Case of the Urease, a Nickel Enzyme.
Chemistry, 25, 2019
|
|
6AT8
 
 | |
6CXT
 
 | | Crystal structure of FAD-dependent dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase fold protein, Butyryl-CoA dehydrogenase, ... | | Authors: | Agarwal, V. | | Deposit date: | 2018-04-04 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Thiotemplated Pyrrole Biosynthesis Gained from the Crystal Structure of Flavin-Dependent Oxidase in Complex with Carrier Protein.
Biochemistry, 58, 2019
|
|
