7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYK
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 5(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYU
 
 | | Structure of the delta dII IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYT
 
 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYH
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYV
 
 | | Structure of the wt IRES eIF5B-containing pre-48S initiation complex, open conformation. Structure 14(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-20 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7TRK
 
 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the agonist iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Belousoff, M.J, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2022-01-29 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor
Elife, 2023
|
|
7TRQ
 
 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the agonist iperoxo and positive allosteric modulator VU0467154 | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, 5-amino-3,4-dimethyl-N-{[4-(trifluoromethanesulfonyl)phenyl]methyl}thieno[2,3-c]pyridazine-6-carboxamide, Antibody fragment scFv16, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Belousoff, M.J, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2022-01-30 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor
Elife, 2023
|
|
7U9Y
 
 | |
7UAS
 
 | |
6UH1
 
 | | Structure of the EVA71 strain 11316 capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
7T1Z
 
 | | Structure of the Fbw7-Skp1-MycNdegron complex | | Descriptor: | F-box/WD repeat-containing protein 7, Myc proto-oncogene N terminal degron, S-phase kinase-associated protein 1, ... | | Authors: | Wang, B, Rusnac, D.V, Clurman, B.E, Zheng, N. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor.
Sci Adv, 8, 2022
|
|
7T1Y
 
 | | Structure of the Fbw7-Skp1-MycCdegron complex | | Descriptor: | F-box/WD repeat-containing protein 7, Myc proto-oncogene protein C terminal degron, S-phase kinase-associated protein 1, ... | | Authors: | Wang, B, Rusnac, D.V, Clurman, B.E, Zheng, N. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two diphosphorylated degrons control c-Myc degradation by the Fbw7 tumor suppressor.
Sci Adv, 8, 2022
|
|
7TUZ
 
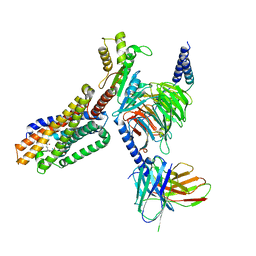 | | Cryo-EM structure of 7alpha,25-dihydroxycholesterol-bound EBI2/GPR183 in complex with Gi protein | | Descriptor: | (2S,4aS,4bS,7R,8S,8aS,9R,10aR)-7-[(2R,3R)-7-hydroxy-3,7-dimethyloctan-2-yl]-4a,7,8-trimethyltetradecahydrophenanthrene-2,9-diol, G-protein coupled receptor 183, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, H, Hung, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
7UCJ
 
 | | Mammalian 80S translation initiation complex with mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
7UCK
 
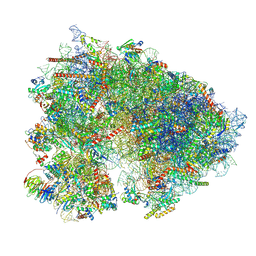 | | 80S translation initiation complex with ac4c(-1) mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
7UNG
 
 | | 48-nm repeat of the human respiratory doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 161, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 45, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-11 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3F3C
 
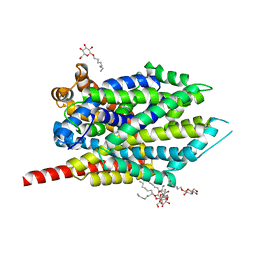 | | Crystal structure of LeuT bound to 4-Fluoro-L-Phenylalanine and sodium | | Descriptor: | 4-FLUORO-L-PHENYLALANINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
7UD5
 
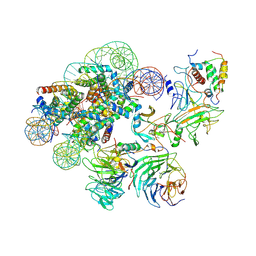 | | Complex between MLL1-WRAD and an H2B-ubiquitinated nucleosome | | Descriptor: | 601 DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Niklas, H.A, Rahman, S, Worden, E.J, Wolberger, C. | | Deposit date: | 2022-03-18 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UYF
 
 | | Human PRMT5:MEP50 structure with Fragment 4 and MTA Bound | | Descriptor: | 4-methyl-1,5-naphthyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7UY1
 
 | | HUMAN PRMT5:MEP50 COMPLEX WITH MTA and Fragment 5 Bound | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-1,5-naphthyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Gunn, R.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7UHY
 
 | | Human GATOR2 complex | | Descriptor: | GATOR complex protein MIOS, GATOR complex protein WDR24, GATOR complex protein WDR59, ... | | Authors: | Rogala, K.B, Valenstein, M.L, Lalgudi, P.V. | | Deposit date: | 2022-03-27 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of the nutrient-sensing hub GATOR2.
Nature, 607, 2022
|
|
3GD9
 
 | | Crystal structure of laminaripentaose-producing beta-1,3-glucanase in complex with laminaritetraose | | Descriptor: | Laminaripentaose-producing beta-1,3-guluase (LPHase), beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Wu, H.M, Hsu, M.T, Liu, S.W, Lai, C.C, Li, Y.K, Wang, W.C. | | Deposit date: | 2009-02-23 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase.
J.Biol.Chem., 284, 2009
|
|
3GD0
 
 | | Crystal structure of laminaripentaose-producing beta-1,3-glucanase | | Descriptor: | Laminaripentaose-producing beta-1,3-guluase (LPHase) | | Authors: | Wu, H.M, Hsu, M.T, Liu, S.W, Lai, C.C, Li, Y.K, Wang, W.C. | | Deposit date: | 2009-02-23 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase.
J.Biol.Chem., 284, 2009
|
|
