3AQU
 
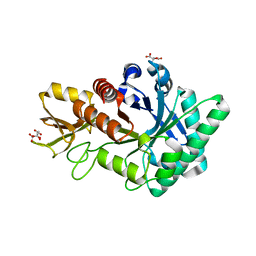 | | Crystal structure of a class V chitinase from Arabidopsis thaliana | | Descriptor: | At4g19810, CITRATE ANION | | Authors: | Numata, T, Ohnuma, T, Osawa, T, Fukamizo, T. | | Deposit date: | 2010-11-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A class V chitinase from Arabidopsis thaliana: gene responses, enzymatic properties, and crystallographic analysis
Planta, 234, 2011
|
|
3TT9
 
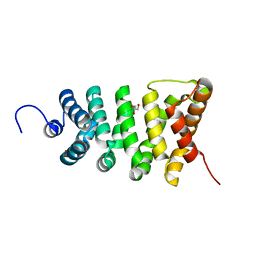 | | Crystal structure of the stable degradation fragment of human plakophilin 2 isoform a (PKP2a) C752R variant | | Descriptor: | GLYCEROL, Plakophilin-2 | | Authors: | Schuetz, A, Roske, Y, Gerull, B, Heinemann, U. | | Deposit date: | 2011-09-14 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations.
Circ Cardiovasc Genet, 5, 2012
|
|
7B9H
 
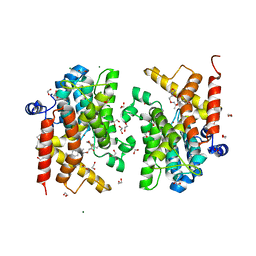 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-42a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ... | | Authors: | Torretta, A, Abbate, S, Parisini, E. | | Deposit date: | 2020-12-14 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
7POZ
 
 | |
3TZ2
 
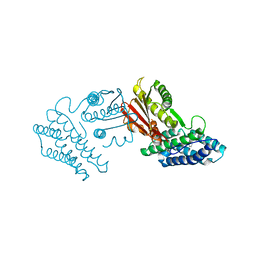 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/phenylbutyrate complex | | Descriptor: | 4-PHENYL-BUTANOIC ACID, [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase, mitochondrial | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2011-09-26 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TJN
 
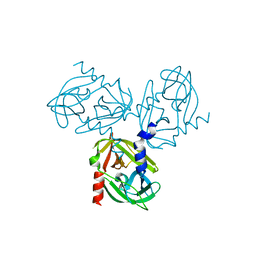 | | HtrA1 catalytic domain, apo form | | Descriptor: | Serine protease HTRA1 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of HtrA1 and Its Subdomains.
Structure, 20, 2012
|
|
3KEC
 
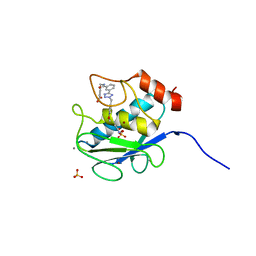 | | Crystal Structure of Human MMP-13 complexed with a phenyl-2H-tetrazole compound | | Descriptor: | 4-{[({3-[2-(4-methoxybenzyl)-2H-tetrazol-5-yl]phenyl}carbonyl)amino]methyl}benzoic acid, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Pavlovsky, A.G, Collins, B, Schnute, M.E. | | Deposit date: | 2009-10-25 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3AX4
 
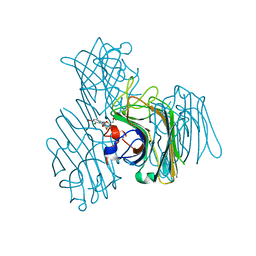 | | Three-dimensional structure of lectin from Dioclea violacea and comparative vasorelaxant effects with Dioclea rostrata | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Bezerra, M.J.B, Bezerra, G.A, Martins, J.L, Nascimento, K.S, Nagano, C.S, Gruber, K, Assereuy, A.M, Delatorre, P, Rocha, B.A.M, Cavada, B.S. | | Deposit date: | 2011-03-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Crystal structure of Dioclea violacea lectin and a comparative study of vasorelaxant properties with Dioclea rostrata lectin
Int.J.Biochem.Cell Biol., 45, 2013
|
|
3L16
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
4ELT
 
 | | Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5-[(4-ethynylphenyl)ethynyl]uridine 5'-(tetrahydrogen triphosphate), ACETATE ION, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of non-polar and "Click-able" nucleotides in the confines of a DNA polymerase active site.
Chem.Commun.(Camb.), 48, 2012
|
|
7PB2
 
 | | Crystal structure of JDI TCR in complex with HLA-A*11:01 bound to KRAS G12D peptide (VVVGADGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS G12D peptide (VVVGADGVGK), MHC class I antigen, ... | | Authors: | Coles, C.H, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Therapeutic high affinity T cell receptor targeting a KRASG12D cancer neoantigen
Nat Commun, 13, 2022
|
|
3AZU
 
 | |
7PC1
 
 | |
3KQE
 
 | | Factor xa in complex with the inhibitor 3-methyl-1-(3-(5- oxo-4,5-dihydro-1h-1,2,4-triazol-3-yl)phenyl)-6-(2'- (pyrrolidin-1-ylmethyl)biphenyl-4-yl)-5,6-dihydro-1h- pyrazolo[3,4-c]pyridin-7(4h)-one | | Descriptor: | 3-METHYL-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-6-(2'-(PYRROLIDIN-1-YLMETHYL)BIPHENYL-4-YL)-5,6-DIHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN-7(4H)-ONE, SODIUM ION, factor Xa heavy chain, ... | | Authors: | Sheriff, S. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4E5R
 
 | | Crystal Structure of Frog DGCR8 Dimerization Domain | | Descriptor: | MGC78846 protein | | Authors: | Guo, F, Senturia, R, Laganowsky, A, Barr, I, Scheidemantle, B.D. | | Deposit date: | 2012-03-14 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dimerization and heme binding are conserved in amphibian and starfish homologues of the microRNA processing protein DGCR8.
Plos One, 7, 2012
|
|
4XTA
 
 | |
4EG0
 
 | |
3U7J
 
 | |
7PXO
 
 | | Structure of the Diels Alderase enzyme AbyU, from Micromonospora maris, co-crystallised with a non transformable substrate analogue | | Descriptor: | (2~{S},4~{S})-1-(4-methoxy-5-methyl-2-oxidanylidene-3~{H}-furan-3-yl)-2,4-dimethyl-dodecane-1,5-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, YD repeat-containing protein | | Authors: | Back, C.R, Race, P.R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Delineation of the Complete Reaction Cycle of a Natural Diels-Alderase
To Be Published
|
|
3KQC
 
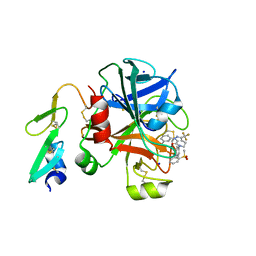 | | Factor xa in complex with the inhibitor 6-(2'- (methylsulfonyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h- 1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-5,6- dihydro-1h-pyrazolo[3,4-c]pyridin-7(4h)-one | | Descriptor: | 6-(2'-(METHYLSULFONYL)BIPHENYL-4-YL)-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-3-(TRIFLUOROMETHYL)-5,6-DIHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN-7(4H)-ONE, SODIUM ION, factor Xa heavy chain, ... | | Authors: | Sheriff, S. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3TL5
 
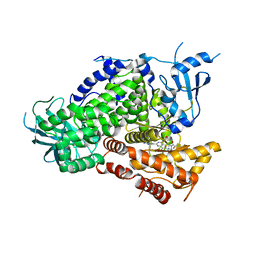 | | Discovery of GDC-0980: a Potent, Selective, and Orally Available Class I Phosphatidylinositol 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Kinase Inhibitor for the Treatment of Cancer | | Descriptor: | (2S)-1-(4-{[2-(2-aminopyrimidin-5-yl)-7-methyl-4-(morpholin-4-yl)thieno[3,2-d]pyrimidin-6-yl]methyl}piperazin-1-yl)-2-hydroxypropan-1-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2011-08-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available Class I Phosphatidylinositol 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Kinase Inhibitor (GDC-0980) for the Treatment of Cancer.
J.Med.Chem., 54, 2011
|
|
4ELU
 
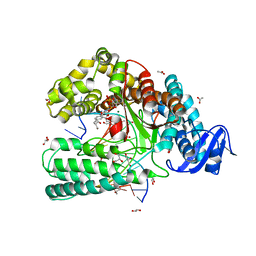 | | Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines | | Descriptor: | 2'-deoxy-5-[(4-ethynylphenyl)ethynyl]cytidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of non-polar and "Click-able" nucleotides in the confines of a DNA polymerase active site.
Chem.Commun.(Camb.), 48, 2012
|
|
3L13
 
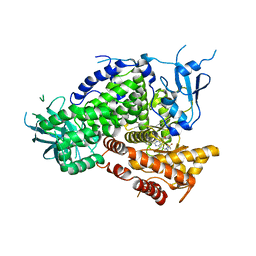 | | Crystal Structures of Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors | | Descriptor: | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, [3-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenyl]methanol | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3L17
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 4-methyl-5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyrimidin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
4EWG
 
 | |
