2WPH
 
 | | factor IXa superactive triple mutant | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2WPK
 
 | | factor IXa superactive triple mutant, ethylene glycol-soaked | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2XYA
 
 | | Non-covalent inhibtors of rhinovirus 3C protease. | | Descriptor: | 2-PHENYLQUINOLIN-4-OL, PICORNAIN 3C | | Authors: | Petersen, J, Edman, K, Edfeldt, F, Johansson, C. | | Deposit date: | 2010-11-16 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-Covalent Inhibitors of Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2WPJ
 
 | | factor IXa superactive triple mutant, NaCl-soaked | | Descriptor: | CALCIUM ION, COAGULATION FACTOR IXA HEAVY CHAIN, COAGULATION FACTOR IXA LIGHT CHAIN, ... | | Authors: | Zogg, T, Brandstetter, H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Cofactor- and Substrate-Assisted Activation of Human Coagulation Factor Ixa
Structure, 17, 2009
|
|
2HXM
 
 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
2XBO
 
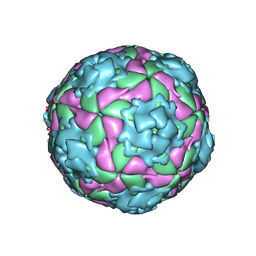 | | Equine Rhinitis A Virus in Complex with its Sialic Acid Receptor | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, P1 | | Authors: | Fry, E.E, Tuthill, T.J, Harlos, K, Walter, T.S, Rowlands, D.J, Stuart, D.I. | | Deposit date: | 2010-04-14 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of Equine Rhinitis a Virus in Complex with its Sialic Acid Receptor.
J.Gen.Virol., 91, 2010
|
|
2XSC
 
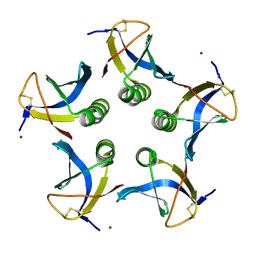 | | Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli | | Descriptor: | SHIGA-LIKE TOXIN 1 SUBUNIT B, ZINC ION | | Authors: | Stein, P.E, Boodhoo, A, Tyrrell, G.J, Brunton, J.L, Oeffner, R.D, Bunkoczi, G, Read, R.J. | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal Structure of the Cell-Binding B Oligomer of Verotoxin-1 from E. Coli.
Nature, 355, 1992
|
|
2XOI
 
 | | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A | | Descriptor: | (2S,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-N-[[(2S,3S,4R,5R)-5-(2,4-DIOXOPYRIMIDIN-1-YL)-4-HYDROXY-2-(HYDROXYMETHYL)OXOLAN-3-YL]METHYLSULFONYL]-3,4-DIHYDROXY-OXOLANE-2-CARBOXAMIDE, RIBONUCLEASE PANCREATIC | | Authors: | Thiyagarajan, N, Smith, B.D, Raines, R.T, Acharya, K.R. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A.
FEBS J., 278, 2011
|
|
2XOG
 
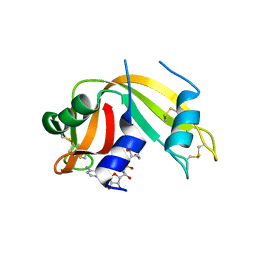 | | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A | | Descriptor: | (2S,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-N-[[(2S,3S,4R,5R)-5-(2,4-DIOXOPYRIMIDIN-1-YL)-4-HYDROXY-2-(HYDROXYMETHYL)OXOLAN-3-YL]METHYLSULFONYL]-3,4-DIHYDROXY-OXOLANE-2-CARBOXAMIDE, RIBONUCLEASE PANCREATIC | | Authors: | Thiyagarajan, N, Smith, B.D, Raines, R.T, Acharya, K.R. | | Deposit date: | 2010-08-16 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A.
FEBS J., 278, 2011
|
|
2EZQ
 
 | |
2G8G
 
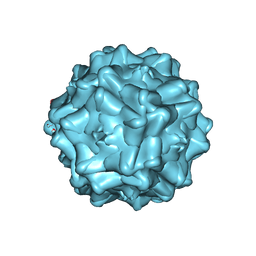 | | Structurally mapping the diverse phenotype of Adeno-Associated Virus serotype 4 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid | | Authors: | Govindasamy, L, Padron, E, McKenna, R, Muzyczka, N, Chiorini, J.A, Agbandje-McKenna, M. | | Deposit date: | 2006-03-02 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally mapping the diverse phenotype of adeno-associated virus serotype 4.
J.Virol., 80, 2006
|
|
2XA8
 
 | | Crystal structure of the Fab domain of omalizumab at 2.41A | | Descriptor: | OMALIZUMAB HEAVY CHAIN, OMALIZUMAB LIGHT CHAIN | | Authors: | Huang, C.H, Hung, F.H.A, Lim, C, Chang, T.W, Ma, C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Physical Basis for Anti-IgE Therapy.
Sci Rep, 5, 2015
|
|
2YGU
 
 | | Crystal structure of fire ant venom allergen, Sol I 2 | | Descriptor: | HEPTANE, VENOM ALLERGEN 2 | | Authors: | Borer, A.S, Wassmann, P, Schirmer, T, Markovic-Housley, Z. | | Deposit date: | 2011-04-20 | | Release date: | 2011-11-23 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Sol I 2, a Major Allergen from Fire Ant Venom
J.Mol.Biol., 415, 2012
|
|
2EZP
 
 | |
2EZS
 
 | |
2H6O
 
 | | Epstein Barr Virus Major Envelope Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major outer envelope glycoprotein gp350, ... | | Authors: | Chen, X.S. | | Deposit date: | 2006-05-31 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Epstein-Barr virus major envelope glycoprotein
Nat.Struct.Mol.Biol., 13, 2006
|
|
2K48
 
 | | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Wang, Y, Boudreaux, D.M, Estrada, D.F, Egan, C.W, St Jeor, S.C, De Guzman, R.N. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein.
J.Biol.Chem., 283, 2008
|
|
2JYW
 
 | | Solution structure of C-terminal domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Chen, K, Harjes, E, Gross, P.J, Fahmy, A, Lu, Y, Shindo, K, Harris, R.S, Matsuo, H. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
Nature, 452, 2008
|
|
3BJL
 
 | |
6QV1
 
 | | Structure of ATPgS-bound outward-facing TM287/288 in complex with nanobody Nb_TM1 | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
6QV2
 
 | | Structure of ATPgS-bound outward-facing TM287/288 in complex with nanobody Nb_TM#2 | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (4.23 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
6QNW
 
 | |
6QPF
 
 | | Influenza A virus Polymerase Heterotrimer A/duck/Fujian/01/2002(H5N1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.634 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QWL
 
 | | Influenza B virus (B/Panama/45) polymerase Hetermotrimer in complex with 3'5' cRNA promoter | | Descriptor: | 3' cRNA, 5' cRNA, Polymerase acidic protein, ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QTJ
 
 | | Crystal structure of human CDK8/CYCC in complex with BI 919811 | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, ~{N},~{N}-dimethyl-2-[4-[4-(2,6-naphthyridin-4-yl)phenyl]pyrazol-1-yl]ethanamide | | Authors: | Boettcher, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
