6JPT
 
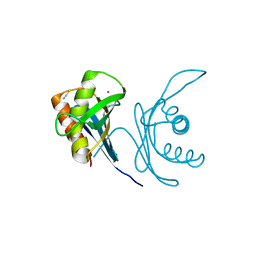 | | Crystal structure of human PAC3 homodimer (trigonal form) | | Descriptor: | POTASSIUM ION, Proteasome assembly chaperone 3, THIOCYANATE ION | | Authors: | Satoh, T, Yagi-Utsumi, M, Okamoto, K, Kurimoto, E, Tanaka, K, Kato, K. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Molecular and Structural Basis of the Proteasome alpha Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer.
Int J Mol Sci, 20, 2019
|
|
5ZJ7
 
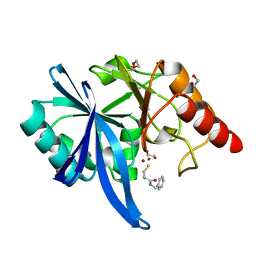 | |
5JUG
 
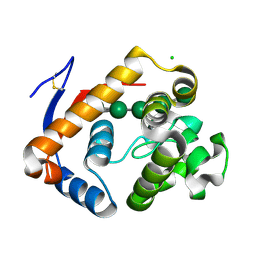 | | Structure of an inactive (E45Q) variant of a beta-1,4-mannanase, SsGH134, in complex with Man5 | | Descriptor: | CHLORIDE ION, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
3ZZP
 
 | | Circular permutant of ribosomal protein S6, lacking edge strand beta- 2 of wild-type S6. | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Saraboji, K, Haglund, E, Lindberg, M.O, Oliveberg, M, Logan, D.T. | | Deposit date: | 2011-09-02 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Trimming Down a Protein Structure to its Bare Foldons: Spatial Organization of the Cooperative Unit.
J.Biol.Chem., 287, 2012
|
|
1UFY
 
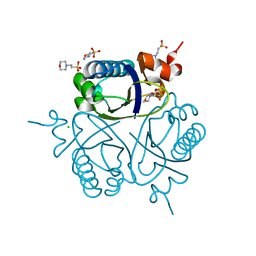 | |
7X4J
 
 | | The 0.96 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with arachidic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The 0.96 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with arachidic acid
To Be Published
|
|
5ZJ1
 
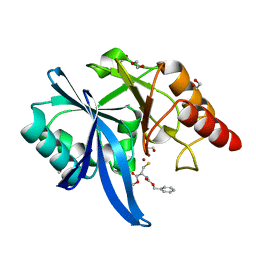 | |
5ZJ8
 
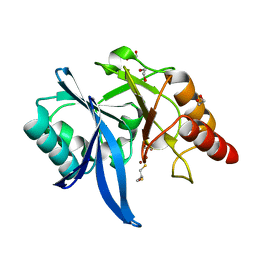 | |
3AGN
 
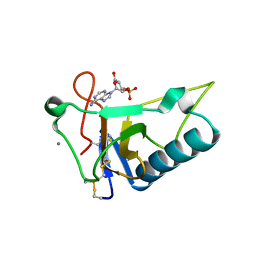 | | Crystal Structure of Ustilago sphaerogena Ribonuclease U2 Complexed with adenosine 3'-monophosphate | | Descriptor: | CALCIUM ION, Ribonuclease U2, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Noguchi, S. | | Deposit date: | 2010-04-03 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Isomerization mechanism of aspartate to isoaspartate implied by structures of Ustilago sphaerogena ribonuclease U2 complexed with adenosine 3'-monophosphate
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3DW5
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
1LUQ
 
 | |
4F18
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 8.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
2E4T
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2006-12-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
3U89
 
 | | Crystal structure of one turn of g/c rich b-dna revisited | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G) -3', MAGNESIUM ION | | Authors: | Maehigashi, T, Woods, K.K, Moulaei, T, Komeda, S, Williams, L.D. | | Deposit date: | 2011-10-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | B-DNA structure is intrinsically polymorphic: even at the level of base pair positions.
Nucleic Acids Res., 40, 2012
|
|
5R2O
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 12, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.969 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6RFH
 
 | |
4M9V
 
 | | Zfp57 mutant (E182Q) in complex with 5-carboxylcytosine DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.969 Å) | | Cite: | DNA recognition of 5-carboxylcytosine by a zfp57 mutant at an atomic resolution of 0.97 angstrom.
Biochemistry, 52, 2013
|
|
5R2Z
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 23, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.969 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
8GEW
 
 | | H-FABP crystal soaked in a bromo palmitic acid solution | | Descriptor: | 2-Bromopalmitic acid, Fatty acid-binding protein, heart, ... | | Authors: | Howard, E, Cousido-Siah, A, Alvarez, A, Espinosa, Y, Podjarny, A, Mitschler, A, Carlevaro, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lipid exchange in crystal-confined fatty acid binding proteins: X-ray evidence and molecular dynamics explanation.
Proteins, 91, 2023
|
|
6HMQ
 
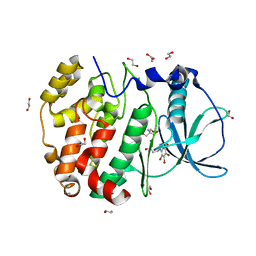 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BENZOTRIAZOLE-TYPE INHIBITOR MB002 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
8PGP
 
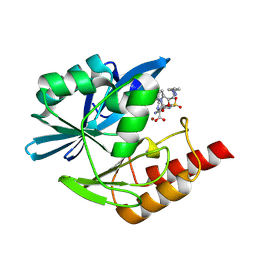 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3193 | | Descriptor: | 7-[(1~{S})-1-[5-[(3-azanylazetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-cyano-4-(methylsulfonylamino)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3193
To Be Published
|
|
7FYV
 
 | | Crystal Structure of human FABP4 in complex with 5-(4-chlorophenyl)-9-methyl-3-oxa-4-azatricyclo[5.2.1.02,6]dec-4-ene-8-carboxylic acid | | Descriptor: | (3aR,4R,5R,6S,7R,7aR)-3-(4-chlorophenyl)-6-methyl-3a,4,5,6,7,7a-hexahydro-4,7-methano-1,2-benzoxazole-5-carboxylic acid, 1,2-ETHANEDIOL, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Wickens, J, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
1M1Q
 
 | | P222 oxidized structure of the tetraheme cytochrome c from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
8B5M
 
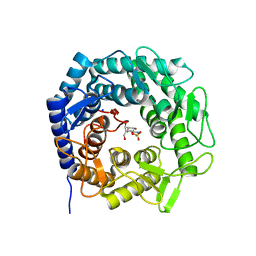 | | Crystal structure of GH47 alpha-1,2-mannosidase from Caulobacter K31 strain in complex with cyclosulfamidate inhibitor | | Descriptor: | (3aR,4S,5S,6R,7R,7aS)-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3a,4,5,6,7,7a-hexahydro-3H-benzo[d][1,2,3]oxathiazole-4,5,6-triol, CALCIUM ION, Mannosyl-oligosaccharide 1,2-alpha-mannosidase, ... | | Authors: | Males, A, Davies, G.J. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | GH47 and Cyclosulfamidate
To Be Published
|
|
3EO6
 
 | |
