4V03
 
 | | MinD cell division protein, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SITE-DETERMINING PROTEIN | | Authors: | Trambaiolo, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
4V02
 
 | | MinC:MinD cell division protein complex, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROBABLE SEPTUM SITE-DETERMINING PROTEIN MINC, ... | | Authors: | Ghosal, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
4WEO
 
 | |
4WFD
 
 | | Structure of the Rrp6-Rrp47-Mtr4 interaction | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex exonuclease RRP6, Exosome complex protein LRP1, ... | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4D5L
 
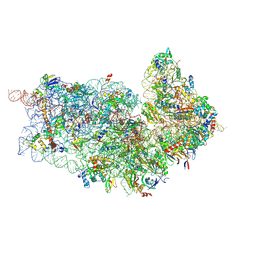 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA 2, 40S RIBOSOMAL PROTEIN ES1, 40S RIBOSOMAL PROTEIN ES10, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D61
 
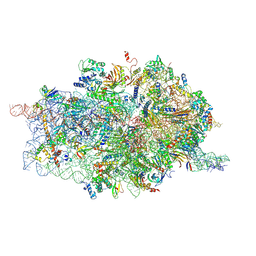 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4RTC
 
 | | Crystal structure of the green fluorescent variant, nowGFP, of the cyan Cerulean at pH 9.0 | | Descriptor: | GLYCEROL, nowGFP | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the green fluorescent protein NowGFP with an anionic tryptophan-based chromophore.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RVR
 
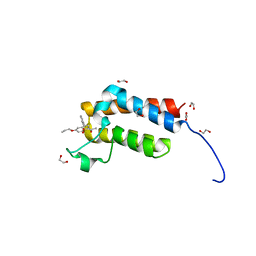 | | Crystal Structure of the bromodomain of human BAZ2B in complex WITH GSK2801 | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(methylsulfonyl)phenyl]-7-propoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-11-27 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
4X8H
 
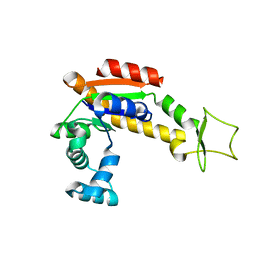 | | Crystal structure of E. coli Adenylate kinase P177A mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8L
 
 | | Crystal structure of E. coli Adenylate kinase P177A mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8O
 
 | | Crystal structure of E. coli Adenylate kinase Y171W mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8M
 
 | | Crystal structure of E. coli Adenylate kinase Y171W mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4XBU
 
 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | Descriptor: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XBR
 
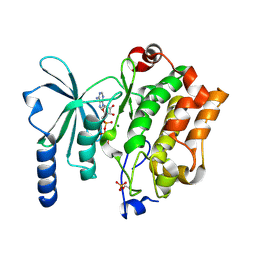 | | In cellulo Crystal Structure of PAK4 in complex with Inka | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein FAM212A,Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XBI
 
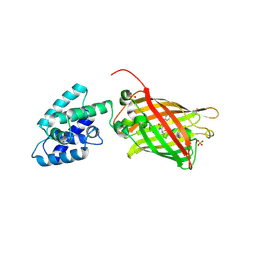 | | Structure Of A Malarial Protein Involved in Proteostasis | | Descriptor: | ClpB protein, putative,Green fluorescent protein, SULFATE ION | | Authors: | Egea, P.F, Ah Young, A.P, Cascio, D. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural mapping of the ClpB ATPases of Plasmodium falciparum: Targeting protein folding and secretion for antimalarial drug design.
Protein Sci., 24, 2015
|
|
4RYS
 
 | |
4RYW
 
 | |
4UER
 
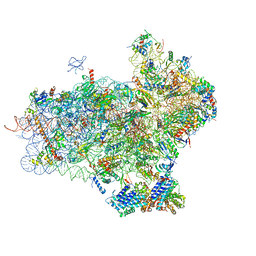 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
3X2S
 
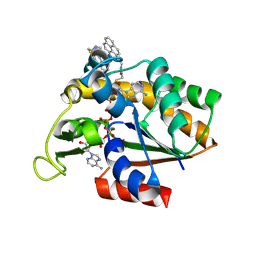 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
4XGY
 
 | | GFP based antibody (fluorobody) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Green fluorescent protein, mAb LCDR3 | | Authors: | Shi, N, Chen, Y.G, Wang, S.H. | | Deposit date: | 2015-01-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | The structure of a GFP-based antibody (fluorobody) to TLH, a toxin from Vibrio parahaemolyticus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XL5
 
 | | X-ray structure of bGFP-A / EGFP complex | | Descriptor: | Green fluorescent protein, bGFP-A | | Authors: | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
4XOV
 
 | |
4XOW
 
 | |
4XVP
 
 | | X-ray structure of bGFP-C / EGFP complex | | Descriptor: | BGFP-C, Green fluorescent protein | | Authors: | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | Deposit date: | 2015-01-27 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
