6Z1J
 
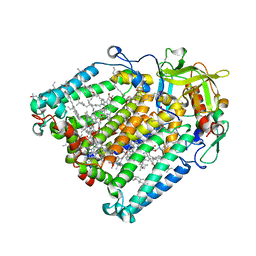 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LSP co-crystallization with spheroidene | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
6ZLA
 
 | |
7BS7
 
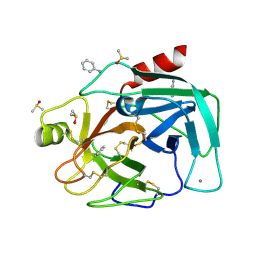 | | Bovine Pancreatic Trypsin with aniline (Cryo) | | Descriptor: | ANILINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
7BS6
 
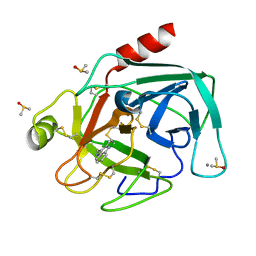 | | Bovine Pancreatic Trypsin with 2-Methyltryptamine (Cryo) | | Descriptor: | 2-(2-methyl-1H-indol-3-yl)ethanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
6ZLK
 
 | | Equilibrium Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid/UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
7BS3
 
 | | Bovine Pancreatic Trypsin with 4-Bromo-benzamidine (Cryo) | | Descriptor: | 4-bromanylbenzenecarboximidamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
8F65
 
 | |
5XG9
 
 | | Crystal Structure of PEG-bound SH3 domain of Myosin IB from Entamoeba histolytica | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, PENTAETHYLENE GLYCOL, ... | | Authors: | Gautam, G, Gourinath, S. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the PEG-bound SH3 domain of myosin IB from Entamoeba histolytica reveals its mode of ligand recognition
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5N5T
 
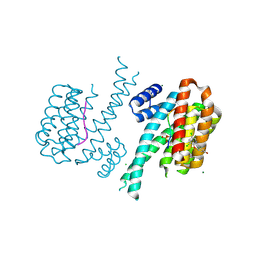 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV2 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyphenol, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
5UG3
 
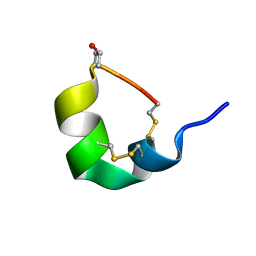 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
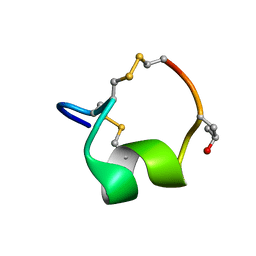 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6CI0
 
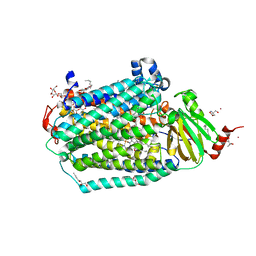 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with E101A (II) mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2018-02-23 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The K-path entrance in cytochrome c oxidase is defined by mutation of E101 and controlled by an adjacent ligand binding domain.
Biochim. Biophys. Acta, 1859, 2018
|
|
5UM3
 
 | |
5D0I
 
 | | Structure of RING finger protein 165 | | Descriptor: | RING finger protein 165, SULFATE ION, ZINC ION | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7YXP
 
 | | Crystal structure of WT AncGR2-LBD WT bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXR
 
 | | Crystal structure of mutant AncGR2-LBD (Y545A) bound to dexamethasone and SHP coregulator fragment | | Descriptor: | Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, FORMIC ACID, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXO
 
 | | Crystal structure of WT AncGR2-LBD bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXN
 
 | | Crystal structure of WT AncGR2-LBD bound to dexamethasone and SHP coregulator fragment | | Descriptor: | Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, FORMIC ACID, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
2IPO
 
 | | E. coli Aspartate Transcarbamoylase complexed with N-phosphonacetyl-L-asparagine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cardia, J.P, Eldo, J, Xia, J, O'Day, E.M, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of L-asparagine and N-phosphonacetyl-L-asparagine to investigate the linkage of catalysis and homotropic cooperativity in E. coli aspartate transcarbomoylase.
Proteins, 71, 2008
|
|
3SPS
 
 | |
6LO9
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 6-cyclohexyloxy-9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86004949 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
4V5X
 
 | | The cryo-EM structure of a 3D DNA-origami object | | Descriptor: | SCAFFOLD STRAND,SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Bai, X.C, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2012-10-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Cryo-Em Structure of a 3D DNA-Origami Object.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5OR9
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 13 | | Descriptor: | (2~{S})-1-(4-fluorophenyl)sulfonyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Vedove, A.D, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
6LOB
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40006661 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
5N5R
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV1 | | Descriptor: | 14-3-3 protein sigma, 2-azanyl-~{N}-(2,6-dimethylphenyl)-~{N}-propan-2-yl-ethanamide, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
