2ACF
 
 | | NMR STRUCTURE OF SARS-COV NON-STRUCTURAL PROTEIN NSP3A (SARS1) FROM SARS CORONAVIRUS | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab | | Authors: | Saikatendu, K.S, Joseph, J.S, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3.
Structure, 13, 2005
|
|
5LBP
 
 | | Oceanobacillus iheyensis macrodomain mutant N30A | | Descriptor: | MacroD-type macrodomain, PHOSPHATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5LW0
 
 | | Oryza sativa APL macrodomain in complex with ADP-ribose | | Descriptor: | Basic helix-loop-helix, putative, expressed, ... | | Authors: | Ariza, A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The role of ADP-ribosylation in regulating DNA interstrand crosslink repair.
J.Cell.Sci., 129, 2016
|
|
5L9Q
 
 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MACROD-TYPE MACRODOMAIN, SULFATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5M3I
 
 | | Macrodomain of Mycobacterium tuberculosis DarG | | Descriptor: | CHLORIDE ION, RNase III inhibitor | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5L9K
 
 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADPR | | Descriptor: | GLYCEROL, MACROD-TYPE MACRODOMAIN, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
6QZU
 
 | | Getah virus macro domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
5LCC
 
 | | Oceanobacillus iheyensis macrodomain mutant D40A | | Descriptor: | MACROD-TYPE MACRODOMAIN | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5IIT
 
 | |
6R0G
 
 | | Getah virus macro domain in complex with ADPr, pose 2 | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural polyprotein | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0P
 
 | | Getah virus macro domain in complex with ADPr in double open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0F
 
 | | Getah virus macro domain in complex with ADPr, pose 1 | | Descriptor: | Non-structural polyprotein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0T
 
 | | Getah virus macro domain in complex with ADPr in open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
5LNC
 
 | |
5ISN
 
 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus | | Descriptor: | Non-structural polyprotein | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Tsika, A.C, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-03-15 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
5IQ5
 
 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5MQX
 
 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5M31
 
 | | Macrodomain of Thermus aquaticus DarG | | Descriptor: | Appr-1-p processing domain protein, CHLORIDE ION, GLYCEROL | | Authors: | Ariza, A. | | Deposit date: | 2016-10-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5KIV
 
 | | Crystal structure of SauMacro (SAV0325) | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Protein-ADP-ribose hydrolase, ... | | Authors: | Williams, R.S, Appel, C.D, Feld, G.K, Wallace, B.D. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the sirtuin-linked macrodomain SAV0325 from Staphylococcus aureus.
Protein Sci., 25, 2016
|
|
5M3E
 
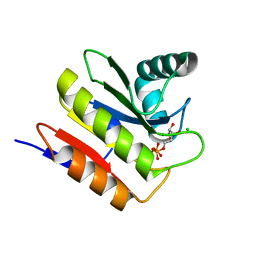 | | Macrodomain of Thermus aquaticus DarG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Appr-1-p processing domain protein, CHLORIDE ION | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
4IQY
 
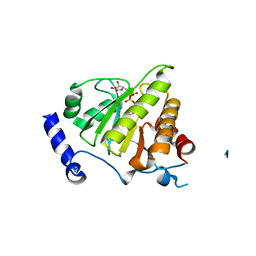 | | Crystal structure of the human protein-proximal ADP-ribosyl-hydrolase MacroD2 | | Descriptor: | MAGNESIUM ION, O-acetyl-ADP-ribose deacetylase MACROD2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Jankevicius, G, Hassler, M, Golia, B, Rybin, V, Zacharias, M, Timinszky, G, Ladurner, A.G. | | Deposit date: | 2013-01-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A family of macrodomain proteins reverses cellular mono-ADP-ribosylation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5AIL
 
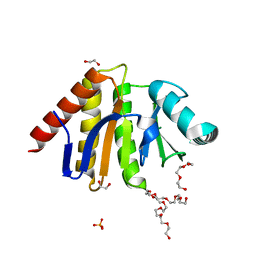 | | Human PARP9 2nd macrodomain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POLY [ADP-RIBOSE] POLYMERASE 9, ... | | Authors: | Sieg, C, Shrestha, L, Talon, R, Sorrell, F, Williams, E, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Elkins, J.M. | | Deposit date: | 2015-02-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Parp9 2Nd Macrodomain
To be Published
|
|
