7D8A
 
 | | Crystal Structure of H3(1-13)/PHF14-PZP fusion protein | | Descriptor: | CALCIUM ION, Gene for histone H3 (germline gene), PHD finger protein 14, ... | | Authors: | Li, H, Zheng, S. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14.
Nucleic Acids Res., 49, 2021
|
|
7D87
 
 | | Crystal Structure of zebrafish PHF14-PZP in complex with H3(1-25) | | Descriptor: | CALCIUM ION, Gene for histone H3 (germline gene), PHD finger protein 14, ... | | Authors: | Li, H, Zheng, S. | | Deposit date: | 2020-10-07 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14.
Nucleic Acids Res., 49, 2021
|
|
5AY8
 
 | | Crystal structure of human nucleosome containing H3.Y | | Descriptor: | CHLORIDE ION, DNA (146-MER), H3.Y, ... | | Authors: | Kujirai, T, Horikoshi, N, Sato, K, Maehara, K, Machida, S, Osakabe, A, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of human histone H3.Y nucleosome
Nucleic Acids Res., 44, 2016
|
|
8W0Q
 
 | | Pembrolizumab CDR-H3 Loop Mimic | | Descriptor: | Pembrolizumab CDR-H3 Loop Mimic | | Authors: | Feig, M, Roche, S.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | De Novo Synthesis and Structural Elucidation of CDR-H3 Loop Mimics.
Acs Chem.Biol., 19, 2024
|
|
4O30
 
 | | Crystal structure of ATXR5 in complex with histone H3.1 and AdoHcy | | Descriptor: | BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase ATXR6, ... | | Authors: | Bergamin, E, Mongeon, V, Couture, J.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication.
Science, 343, 2014
|
|
5VAC
 
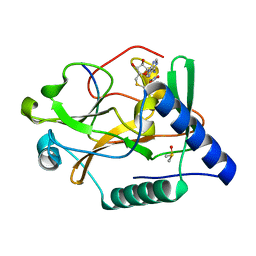 | | Crystal Structure of ATXR5 SET domain in complex with K36me3 histone H3 peptide | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5T8R
 
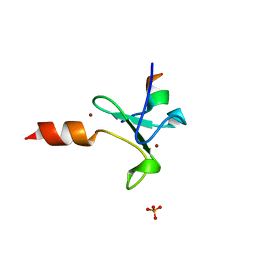 | | Crystal structure of human BAZ2A PHD zinc finger in complex with unmodified H3 10-mer | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, Histone H3.1, ... | | Authors: | Amato, A, Gadd, M.S, Bortoluzzi, A, Ciulli, A. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of molecular recognition of helical histone H3 tail by PHD finger domains.
Biochem. J., 474, 2017
|
|
5VAH
 
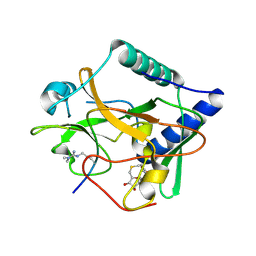 | | Crystal structure of ATXR5 SET domain in complex with histone H3 di-methylated on R26 | | Descriptor: | Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
7W5M
 
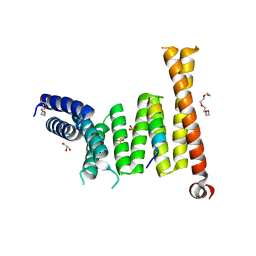 | |
5VBC
 
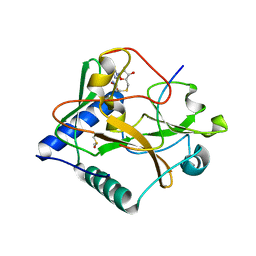 | | Crystal structure of ATXR5 in complex with histone H3.1 | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.1 peptide, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Couture, J.-F, Bergamin, E. | | Deposit date: | 2017-03-29 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
1GUW
 
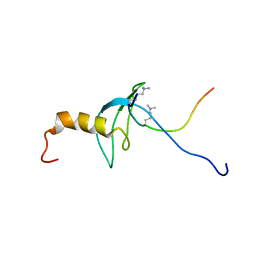 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2025-04-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
4HGA
 
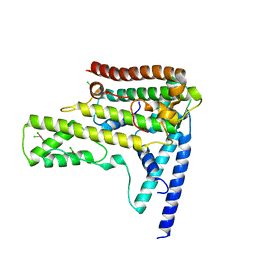 | | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Liu, C.P, Xiong, C.Y, Wang, M.Z, Yu, Z.L, Yang, N, Chen, P, Zhang, Z.G, Li, G.H, Xu, R.M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4N4I
 
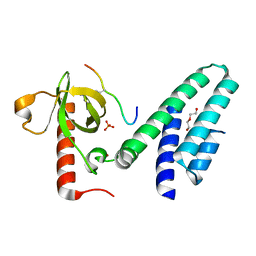 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4H
 
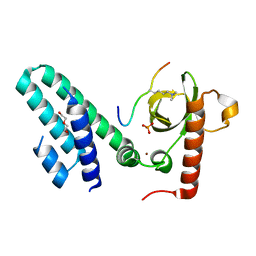 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4ZBJ
 
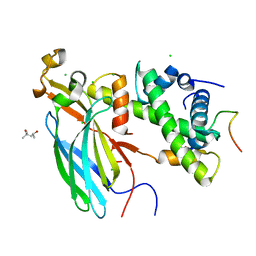 | | UBN1 peptide bound to H3.3/H4/Asf1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Marmorstein, R, Ricketts, M.D, Tang, Y. | | Deposit date: | 2015-04-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex.
Nat Commun, 6, 2015
|
|
2H6Q
 
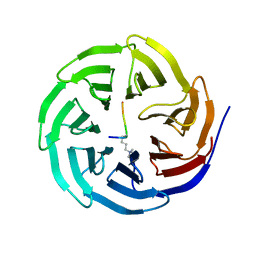 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me3 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3ZVY
 
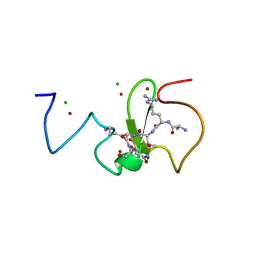 | | PHD finger of human UHRF1 in complex with unmodified histone H3 N- terminal tail | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE UHRF1, ... | | Authors: | Lallous, N, Birck, C, Mc Ewen, A.G, Legrand, P, Samama, J.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phd Finger of Human Uhrf1 Reveals a New Subgroup of Unmethylated Histone H3 Tail Readers.
Plos One, 6, 2011
|
|
5VAB
 
 | | Crystal structure of ATXR5 PHD domain in complex with histone H3 | | Descriptor: | ATXR5 PHD domain, Histone H3 peptide, ZINC ION | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
1N3J
 
 | | Structure and Substrate of a Histone H3 Lysine Methyltransferase from Paramecium Bursaria Chlorella Virus 1 | | Descriptor: | Histone H3 Lysine Methyltransferase | | Authors: | Manzur, K.L, Farooq, A, Zeng, L, Plotnikova, O, Sachchidanand, Koch, A.W, Zhou, M.-M. | | Deposit date: | 2002-10-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3.
Nat.Struct.Biol., 10, 2003
|
|
3WTP
 
 | | Crystal Structure of the heterotypic nucleosome containing human CENP-A and H3.3 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Arimura, Y, Shirayama, K, Horikoshi, N, Fujita, R, Kagawa, W, Fukagawa, T, Almouzni, G, Kurumizaka, H. | | Deposit date: | 2014-04-14 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3
Sci Rep, 4, 2014
|
|
5BSA
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.611 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BO0
 
 | |
4Z5T
 
 | | The nucleosome containing human H3.5 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Urahama, T, Harada, A, Maehara, K, Horikoshi, N, Sato, K, Sato, Y, Shiraishi, K, Sugino, N, Osakabe, A, Tachiwana, H, Kagawa, W, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-04-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis.
Epigenetics Chromatin, 9, 2016
|
|
5FB1
 
 | | Crystal Structure of a PHD finger bound to histone H3 K9me3 peptide | | Descriptor: | MALONATE ION, Nuclear autoantigen Sp-100, Peptide from Histone H3, ... | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
5B0Z
 
 | | The crystal structure of the nucleosome containing H3.2, at 1.98 A resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Suzuki, Y, Horikoshi, N, Kato, D, Kurumizaka, H. | | Deposit date: | 2015-11-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122
Biochem.Biophys.Res.Commun., 469, 2016
|
|
