7AWT
 
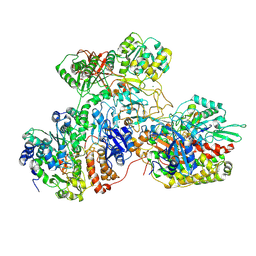 | | E. coli NADH quinone oxidoreductase hydrophilic arm | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schimpf, J, Grishkovskaya, I, Haselbach, D, Friedrich, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of the peripheral arm of a minimalistic respiratory complex I.
Structure, 30, 2022
|
|
2XRY
 
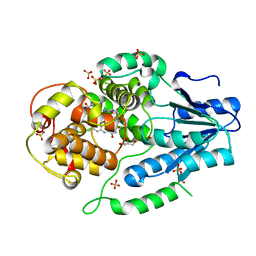 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
2XRZ
 
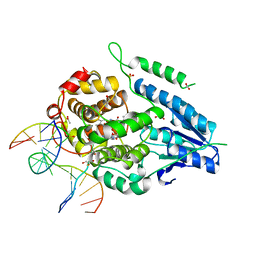 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei in complex with intact CPD-lesion | | Descriptor: | ACETATE ION, COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
2YAK
 
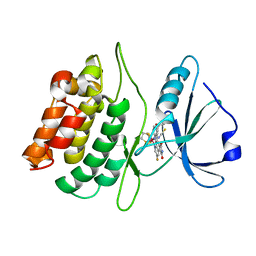 | | Structure of death-associated protein Kinase 1 (dapk1) in complex with a ruthenium octasporine ligand (OSV) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, RUTHENIUM OCTASPORINE 4 | | Authors: | Feng, L, Geisselbrecht, Y, Blanck, S, Wilbuer, A, Atilla-Gokcumen, G.E, Filippakopoulos, P, Kraeling, K, Celik, M.A, Harms, K, Maksimoska, J, Marmorstein, R, Frenking, G, Knapp, S, Essen, L.-O, Meggers, E. | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structurally Sophisticated Octahedral Metal Complexes as Highly Selective Protein Kinase Inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
4LL6
 
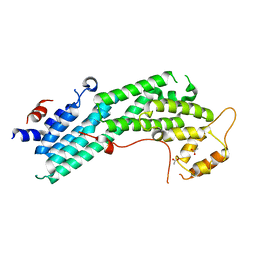 | | Structure of Myo4p globular tail domain. | | Descriptor: | ACETIC ACID, Myosin-4 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3H4X
 
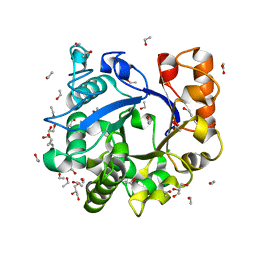 | |
4LL7
 
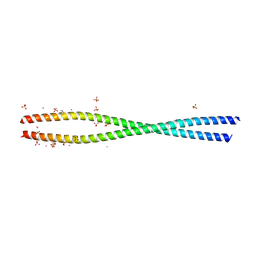 | | Structure of She3p amino terminus. | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DYSPROSIUM ION, ... | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LL8
 
 | | Complex of carboxy terminal domain of Myo4p and She3p middle fragment | | Descriptor: | Myosin-4, SWI5-dependent HO expression protein 3 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.578 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1D1N
 
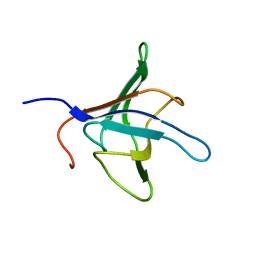 | | SOLUTION STRUCTURE OF THE FMET-TRNAFMET BINDING DOMAIN OF BECILLUS STEAROTHERMOPHILLUS TRANSLATION INITIATION FACTOR IF2 | | Descriptor: | INITIATION FACTOR 2 | | Authors: | Meunier, S, Spurio, S, Czisch, M, Wechselberger, R, Gueunneugues, M. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2.
EMBO J., 19, 2000
|
|
3TV4
 
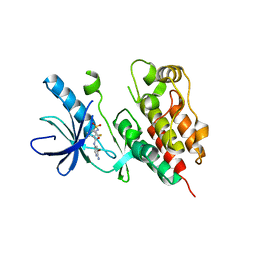 | | Human B-Raf Kinase Domain in Complex with an Bromopyridine Benzamide Inhibitor | | Descriptor: | N-(6-amino-5-bromopyridin-3-yl)-2,6-difluoro-3-[(propylsulfonyl)amino]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Selby, L.T, Wu, W.-I. | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Pyrazolopyridine Inhibitors of B-Raf(V600E). Part 1: The Development of Selective, Orally Bioavailable, and Efficacious Inhibitors.
ACS Med Chem Lett, 2, 2011
|
|
1D9S
 
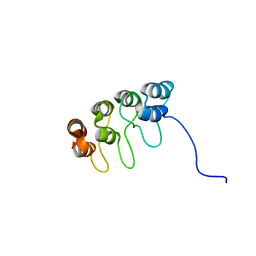 | | TUMOR SUPPRESSOR P15(INK4B) STRUCTURE BY COMPARATIVE MODELING AND NMR DATA | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR B | | Authors: | Yuan, C, Ji, L, Selby, T.L, Byeon, I.J.L, Tsai, M.D. | | Deposit date: | 1999-10-29 | | Release date: | 2000-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: comparisons of conformational properties between p16(INK4A) and p18(INK4C).
J.Mol.Biol., 294, 1999
|
|
2LRD
 
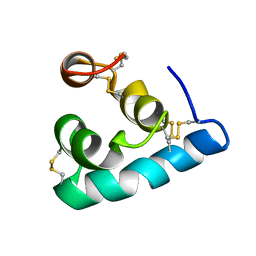 | | The solution structure of the monomeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2NVJ
 
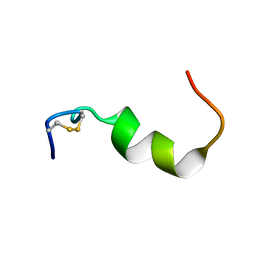 | | NMR structures of transmembrane segment from subunit a from the yeast proton V-ATPase | | Descriptor: | 25mer peptide from Vacuolar ATP synthase subunit a, vacuolar isoform | | Authors: | Hemminga, M.A, van Mierlo, C.P, Wechselberger, R, de Jong, E.R, Duarte, A.M. | | Deposit date: | 2006-11-13 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Segment TM7 from the cytoplasmic hemi-channel from V(O)-H(+)-V-ATPase includes a flexible region that has a potential role in proton translocation
Biochim.Biophys.Acta, 1768, 2007
|
|
4AS0
 
 | | Cyclometalated Phthalimides as Protein Kinase Inhibitors | | Descriptor: | PHTALIMIDE-RUTHENIUM COMPLEX, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Blanck, S, Geisselbrecht, Y, Middel, S, Mietke, T, Harms, K, Essen, L.-O, Meggers, E. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Cyclometalated Phthalimides: Design, Synthesis and Kinase Inhibition.
Dalton Trans, 41, 2012
|
|
2LRE
 
 | | The solution structure of the dimeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
8AMZ
 
 | | Spinach 19S proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1 homolog, 26S proteasome non-ATPase regulatory subunit 2 homolog, 26S proteasome regulatory subunit 7, ... | | Authors: | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the plant 26S proteasome
Plant Communications, 3, 2022
|
|
6U7T
 
 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | O'Shea Murray, V.L, Cao, S, Horvath, M.P, David, S.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
7NKU
 
 | | diazaborine bound Drg1(AFG2) | | Descriptor: | 6-METHYL-2(PROPANE-1-SULFONYL)-2H-THIENO[3,2-D][1,2,3]DIAZABORININ-1-OL, ATPase family gene 2 protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Prattes, M, Bergler, H, Haselbach, D. | | Deposit date: | 2021-02-19 | | Release date: | 2021-06-23 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inhibition of the AAA-ATPase Drg1 by diazaborine.
Nat Commun, 12, 2021
|
|
6OT9
 
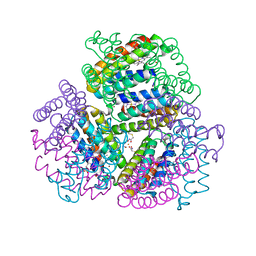 | | Bimetallic dodecameric cage design 1 (BMC1) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OT4
 
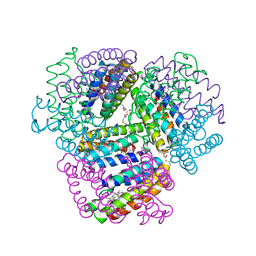 | | Bimetallic dodecameric cage design 2 (BMC2) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OT7
 
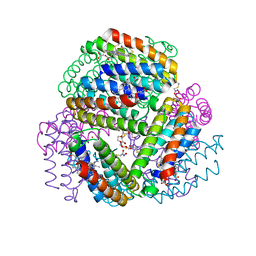 | | Bimetallic dodecameric cage design 3 (BMC3) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OT8
 
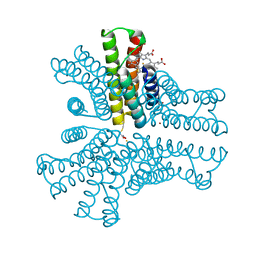 | | Bimetallic hexameric cage design 4 (BMC4) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
7NH3
 
 | | Nematocida Huwe1 in open conformation. | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Petrova, O, Grishkovskaya, I, Grabarczyk, D.B, Kessler, D, Haselbach, D, Clausen, T. | | Deposit date: | 2021-02-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.37 Å) | | Cite: | Crystal structure of HUWE1: One ring to ubiquitinate them all
To Be Published
|
|
8DW7
 
 | | DNA glycosylase MutY variant N146S in complex with DNA containing the transition state analog 1N paired with d(8-oxo-G) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-31 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DVP
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with substrate purine | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
