3BHP
 
 | | Crystal structure of UPF0291 protein ynzC from Bacillus subtilis at resolution 2.0 A. Northeast Structural Genomics Consortium target SR384 | | Descriptor: | UPF0291 protein ynzC | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Janjua, H, Cunningham, K, Maglaqui, M, Owens, L.A, Zhao, L, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the UPF0291 protein ynzC from Bacillus subtilis at the resolution 2.0 A.
To be Published
|
|
5XHV
 
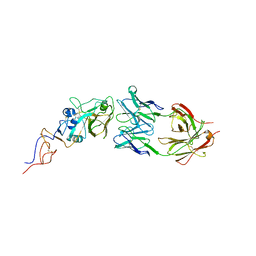 | | Crystal Structure Of Fab S40 In Complex With Influenza Hemagglutinin, HA1 subunit. | | Descriptor: | Hemagglutinin, S40 heavy chain, S40 light chain | | Authors: | Lee, C.C, Wang, A.H.J, Yu, C.M, Yang, A.S. | | Deposit date: | 2017-04-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries.
Sci Rep, 7, 2017
|
|
2J60
 
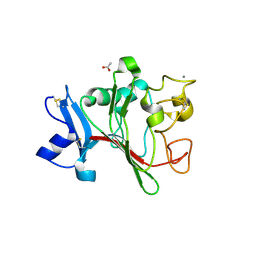 | | H-ficolin complexed to D-fucose | | Descriptor: | ACETATE ION, CALCIUM ION, FICOLIN-3, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-09-21 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
3IRB
 
 | |
2MOA
 
 | |
4CDK
 
 | | Structure of ZNRF3-RSPO1 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3, R-SPONDIN-1 | | Authors: | Peng, W.C, de Lau, W, Madoori, P.K, Forneris, F, Granneman, J.C.M, Clevers, H, Gros, P. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Wnt-Antagonist Znrf3 and its Complex with R-Spondin 1 and Implications for Signaling.
Plos One, 8, 2013
|
|
1MR3
 
 | | Saccharomyces cerevisiae ADP-ribosylation Factor 2 (ScArf2) complexed with GDP-3'P at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ADP-ribosylation factor 2, ... | | Authors: | Amor, J.-C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases.
J.Biol.Chem., 276, 2001
|
|
4C16
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | | Descriptor: | (1R,2R,3S)-3-methylcyclohexane-1,2-diol, (S)-CYCLOHEXYL LACTIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2013-08-09 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
5EFZ
 
 | | Monoclinic structure of the acetyl esterase MekB | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Toelzer, C, Pal, S, Watzlawick, H, Altenbuchner, J, Niefind, K. | | Deposit date: | 2015-10-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
2J0H
 
 | | L-ficolin complexed to acetyl-choline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE, CALCIUM ION, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-03 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
3RKI
 
 | | Structural basis for immunization with post-fusion RSV F to elicit high neutralizing antibody titers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Swanson, K.A, Settembre, E.C, Shaw, C.A, Dey, A.K, Rappuoli, R, Mandl, C.W, Dormitzer, P.D, Carfi, A. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1N3I
 
 | | Crystal Structure of Mycobacterium tuberculosis PNP with transition state analog DADMe-ImmH | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine Nucleoside Phosphorylase | | Authors: | Lewandowicz, A, Shi, W, Evans, G.B, Tyler, P.C, Furneaux, R.H, Basso, L.A, Santos, D.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Over-The-Barrier Transition State Analogues Provide New Chemistries for Inhibitor Design: The Case of Purine Nucleoside Phosphorylase
BIOCHEMISTRY, 42, 2003
|
|
5Q67
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 144) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q6I
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 155) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3WB8
 
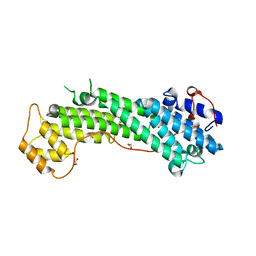 | | Crystal Structure of MyoVa-GTD | | Descriptor: | 1,2-ETHANEDIOL, Unconventional myosin-Va | | Authors: | Wei, Z, Liu, X, Yu, C, Zhang, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LIT
 
 | |
5AW0
 
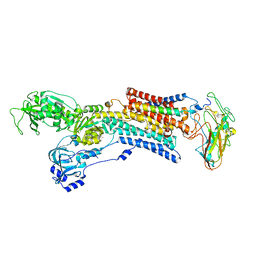 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 55 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
2NZA
 
 | |
2OEF
 
 | |
4LJD
 
 | | Structure of a photobleached state of IrisFP under low intensity laser-light | | Descriptor: | Green to red photoconvertible GPF-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Duan, C, Adam, V, Byrdin, M, Ridard, J, Kieffer-Jacquinod, S, Morlot, C, Arcizet, D, Demachy, I, Bourgeois, D. | | Deposit date: | 2013-07-04 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural evidence for a two-regime photobleaching mechanism in a reversibly switchable fluorescent protein.
J.Am.Chem.Soc., 135, 2013
|
|
1KTB
 
 | | The Structure of alpha-N-Acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Garman, S.C, Hannick, L, Zhu, A, Garboczi, D.N. | | Deposit date: | 2002-01-15 | | Release date: | 2002-03-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of alpha-N-acetylgalactosaminidase: molecular basis of glycosidase deficiency diseases
Structure, 10, 2002
|
|
2OIP
 
 | |
5AVY
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 20 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
1B27
 
 | |
3U1I
 
 | | Dengue virus protease covalently bound to a peptide | | Descriptor: | SULFATE ION, Serine protease NS3, Serine protease subunit NS2B, ... | | Authors: | Noble, C.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand-bound structures of the dengue virus protease reveal the active conformation
J.Virol., 86, 2012
|
|
