7RVF
 
 | |
9GO6
 
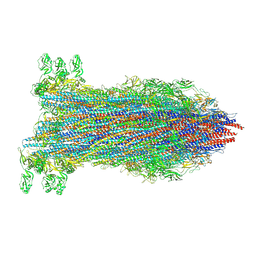 | | Salmonella hook-filament junction complex | | Descriptor: | Flagellar hook protein FlgE, Flagellar hook-associated protein, Flagellar hook-associated protein 1, ... | | Authors: | Qin, K, Einenkel, R, Erhardt, E, Bergeron, J.R. | | Deposit date: | 2024-09-04 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structure of the complete extracellular bacterial flagellum reveals the mechanism of flagellin incorporation.
Nat Microbiol, 10, 2025
|
|
7RRF
 
 | | Carbonic Anhydrase IX-mimic in complex with Beta-Galactose_2C | | Descriptor: | 2-[1-(1,1,3-trioxo-2,3-dihydro-1H-1lambda~6~,2-benzothiazol-6-yl)-1H-1,2,3-triazol-4-yl]ethyl beta-L-gulopyranoside, Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, McKenna, R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of Carbonic Anhydrase IX and Monocarboxylate Transporters 1 and 4 in breast cancer via novel inhibition with Beta-Galactose 2C
To Be Published
|
|
1MAW
 
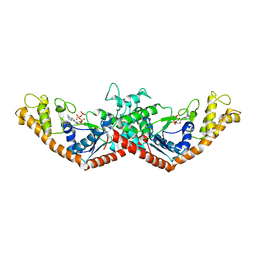 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with ATP in an Open Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, TRYPTOPHAN-TRNA LIGASE | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
9FZ5
 
 | | Cryo-EM structure of LptDE-YedD complex from Escherichia Coli | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, Uncharacterized lipoprotein YedD | | Authors: | Nguyen, V.S, Remaut, H, Collet, J.F, Gennaris, A, Thouvenel, L. | | Deposit date: | 2024-07-04 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Optimal functioning of the Lpt bridge depends on a ternary complex between the lipocalin YedD and the LptDE translocon.
Cell Rep, 44, 2025
|
|
1MK4
 
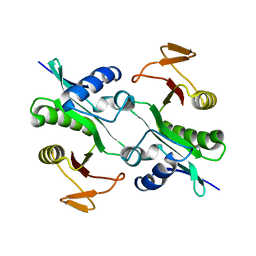 | | Structure of Protein of Unknown Function YqjY from Bacillus subtilis, Probable Acetyltransferase | | Descriptor: | Hypothetical protein yqjY | | Authors: | Zhang, R, Dementiva, I, Mo, A, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A crystal structure of a hypothetical protein yqjY
from Bacillus subtilis
To be Published
|
|
7R80
 
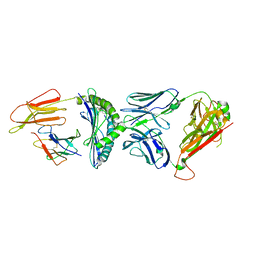 | | Crystal structure of C3 TCR complex with QW9-bound HLA-B*5301 | | Descriptor: | Alpha chain of C3 TCR, Beta Chain of C3 TCR, Beta-2-microglobulin, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
9GB0
 
 | | Extended phiCD508 portal adjacent capsid | | Descriptor: | gp48 - Minor capsid protein, gp49 - Major capsid protein | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
1MKL
 
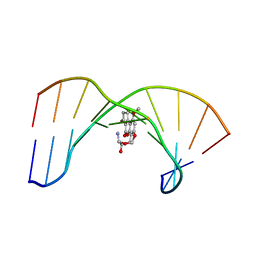 | | NMR REFINED STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT IN A 5'-CPAFBG-3' SEQUENCE | | Descriptor: | 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3', 5'-D(*AP*GP*AP*TP*CP*GP*AP*TP*GP*T)-3', 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 | | Authors: | Giri, I, Jenkins, M.D, Schnetz-Boutaud, N.C, Stone, M.P. | | Deposit date: | 2002-08-29 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural refinement of the 8,9-dihydro-8-(N7-guanyl)-9-hydroxy-aflatoxin B(1) adduct in a 5'-Cp(AFB)G-3' sequence.
Chem.Res.Toxicol., 15, 2002
|
|
1MKR
 
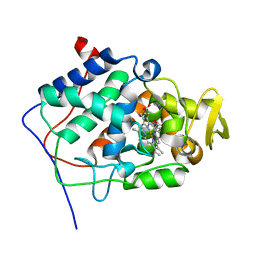 | | Crystal Structure of a Mutant Variant of Cytochrome c Peroxidase (Plate like crystals) | | Descriptor: | Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
7RWP
 
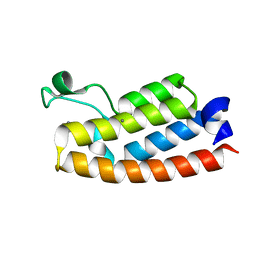 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | Descriptor: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
9G0Z
 
 | |
1ML5
 
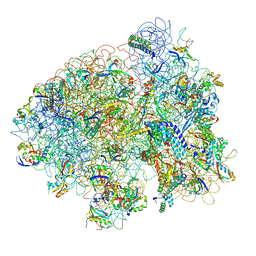 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | Deposit date: | 2002-08-30 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
9G58
 
 | |
7R8T
 
 | |
9GM8
 
 | | MukBEF in a nucleotide-bound state with open neck gate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Burmann, F, Lowe, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic.
Cell, 188, 2025
|
|
1MC1
 
 | | BETA-LACTAM SYNTHETASE WITH PRODUCT (DGPC), AMP AND PPI | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-LACTAM SYNTHETASE, DEOXYGUANIDINOPROCLAVAMINIC ACID, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2002-08-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MCR
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
7RPC
 
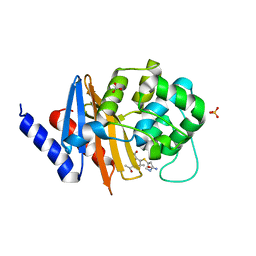 | | X-ray crystal structure of OXA-24/40 K84D in complex with ertapenem | | Descriptor: | (1S,4R,5S,6S)-3-{[(3S,5S)-5-carbamoylpyrrolidin-3-yl]sulfanyl}-6-[(1R)-1-hydroxyethyl]-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
9GLA
 
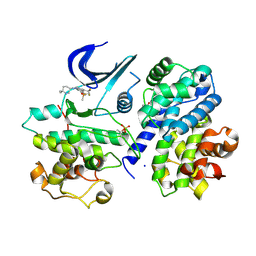 | | Crystal structure of a CDK2-based CDK7 mimic with inhibitor SY5609 | | Descriptor: | 7-dimethylphosphoryl-3-[2-[[(3~{S})-6,6-dimethylpiperidin-3-yl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]-1~{H}-indole-6-carbonitrile, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Skerlova, J, Krejcirikova, V, Rezacova, P. | | Deposit date: | 2024-08-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | CDK2-based CDK7 mimic as a tool for structural analysis: Biochemical validation and crystal structure with SY5609.
Int.J.Biol.Macromol., 294, 2025
|
|
1MD6
 
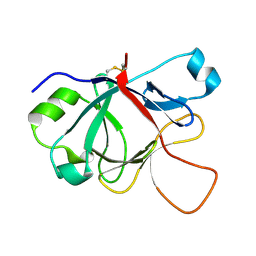 | | High resolution crystal structure of murine IL-1F5 reveals unique loop conformation for specificity | | Descriptor: | interleukin 1 family, member 5 (delta) | | Authors: | Pei, X.Y, Dunn, E.F, Gay, N.J, O'Neill, L.A. | | Deposit date: | 2002-08-07 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Structure of Murine Interleukin 1 Homologue IL-1F5 Reveals Unique Loop Conformations for Receptor Binding Specificity.
Biochemistry, 42, 2003
|
|
9G8S
 
 | | C3 reconstruction of extended phiCD508 needle | | Descriptor: | Baseplate J family protein, FE (II) ION, Peptidoglycan-binding LysM protein, ... | | Authors: | Wilson, J.S, Fagan, R.P, Bullough, P.A. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Molecular mechanism of bacteriophage contraction structure of an S-layer-penetrating bacteriophage.
Life Sci Alliance, 8, 2025
|
|
9G0X
 
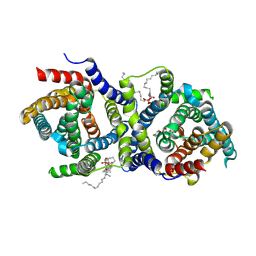 | |
7RUH
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844 | | Descriptor: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | Authors: | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844
To Be Published
|
|
1MI8
 
 | | 2.0 Angstrom crystal structure of a DnaB intein from Synechocystis sp. PCC 6803 | | Descriptor: | DnaB intein | | Authors: | Ding, Y, Chen, X, Ferrandon, S, Xu, M, Rao, Z. | | Deposit date: | 2002-08-22 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing
J.Biol.Chem., 278, 2003
|
|
