3MVX
 
 | | X-ray structure of the reduced NikA/1 hybrid, NikA/1-Red | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2-[2-[carboxymethyl(phenylmethyl)amino]ethyl-[(2-hydroxyphenyl)methyl]amino]ethanoic acid, ... | | Authors: | Cavazza, C, Bochot, C, Rousselot-Pailley, P, Carpentier, P, Cherrier, M.V, Martin, L, Marchi-Delapierre, C, Fontecilla-Camps, J.C, Menage, S. | | Deposit date: | 2010-05-05 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of the reaction of aromatic C-H with O(2) catalysed by a protein-bound iron complex
NAT.CHEM., 2, 2010
|
|
1BSU
 
 | |
3U36
 
 | | Crystal Structure of PG9 Fab | | Descriptor: | PG9 Fab heavy chain, PG9 Fab light chain, SULFATE ION | | Authors: | McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
4QVH
 
 | | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein | | Descriptor: | CITRATE ANION, COENZYME A, GLYCEROL, ... | | Authors: | Jung, J, Bashiri, G, Johnston, J.M, Baker, E.N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein.
J.Struct.Biol., 188, 2014
|
|
3V2Y
 
 | | Crystal Structure of a Lipid G protein-Coupled Receptor at 2.80A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sphingosine 1-phosphate receptor 1, Lysozyme chimera (E.C.3.2.1.17), ... | | Authors: | Hanson, M.A, Roth, C.B, Jo, E, Griffith, M.T, Scott, F.L, Reinhart, G, Desale, H, Clemons, B, Cahalan, S.M, Schuerer, S.C, Sanna, M.G, Han, G.W, Kuhn, P, Rosen, H, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a lipid G protein-coupled receptor.
Science, 335, 2012
|
|
4DAW
 
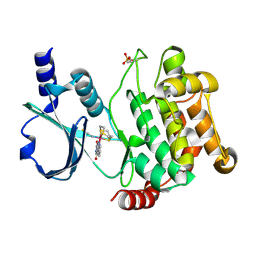 | | Crystal structure of PAK1 kinase domain with the ruthenium phthalimide complex | | Descriptor: | Serine/threonine-protein kinase PAK 1, [1,3-dioxo-6-(pyridin-2-yl-kappaN)-2,3-dihydro-1H-isoindol-5-yl-kappaC~5~][(thioxomethylidene)azanido-kappaN](1,4,7-trithionane-kappa~3~S~1~,S~4~,S~7~)ruthenium | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The art of filling protein pockets efficiently with octahedral metal complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2OQL
 
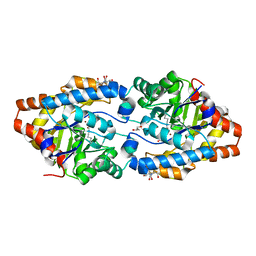 | | Structure of Phosphotriesterase mutant H254Q/H257F | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Parathion hydrolase, ... | | Authors: | Kim, J, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of PTE mutant H254Q/H257F
To be Published
|
|
3MTU
 
 | | Structure of the Tropomyosin Overlap Complex from Chicken Smooth Muscle | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid assembly scaffolding protein,Tropomyosin alpha-1 chain, ... | | Authors: | Klenchin, V.A, Frye, J, Rayment, I. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition .
Biochemistry, 49, 2010
|
|
2Z8E
 
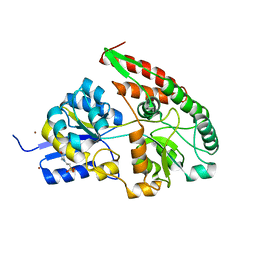 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with galacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
4RE3
 
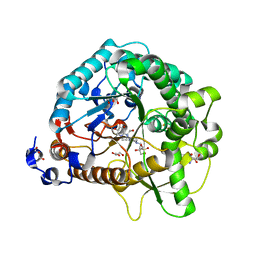 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
4HMD
 
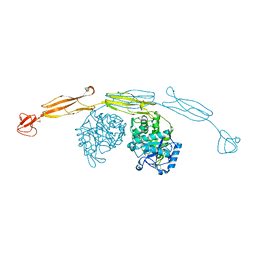 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3CCB
 
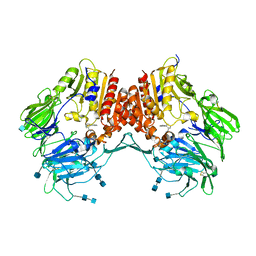 | | Crystal Structure of Human DPP4 in complex with a benzimidazole derivative | | Descriptor: | 1-biphenyl-2-ylmethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wallace, M.B, Skene, R.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based design and synthesis of benzimidazole derivatives as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4MPV
 
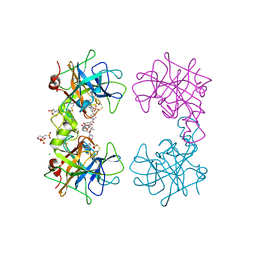 | | Human beta-tryptase co-crystal structure with (2R,4S)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-4-hydroxy-2-(2-hydroxypropan-2-yl)-5,5-dimethyl-1,3-dioxolane-2,4-dicarboxamide | | Descriptor: | (2R,4S)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-4-hydroxy-2-(2-hydroxypropan-2-yl)-5,5-dimethyl-1,3-dioxolane-2,4-dicarboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Target-Directed Self-Assembly of Homodimeric Drugs Against beta-Tryptase.
Acs Med.Chem.Lett., 9, 2018
|
|
2DUV
 
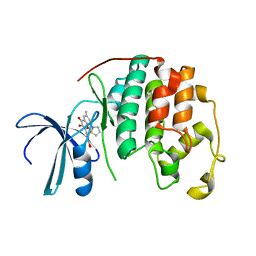 | | Structure of CDK2 with a 3-hydroxychromones | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)-8-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-3-HYDROXY-6-METHYL-4H-CHROMEN-4-ONE, Cell division protein kinase 2 | | Authors: | Kim, K.H, Lee, J, Park, T, Jeong, S, Hong, C. | | Deposit date: | 2006-07-27 | | Release date: | 2007-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Hydroxychromones as cyclin-dependent kinase inhibitors: synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1PT2
 
 | |
2OXB
 
 | | Crystal structure of a cell-wall invertase (E203Q) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2007-02-20 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternate sucrose binding mode in the E203Q Arabidopsis invertase mutant: An X-ray crystallography and docking study.
Proteins, 71, 2007
|
|
1IE7
 
 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
4HME
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2DE8
 
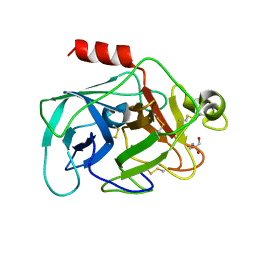 | |
1XXO
 
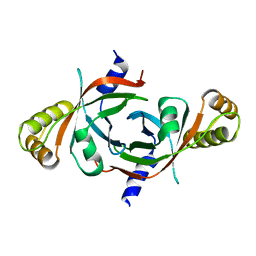 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution | | Descriptor: | hypothetical protein Rv1155 | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution
To be Published
|
|
1XL2
 
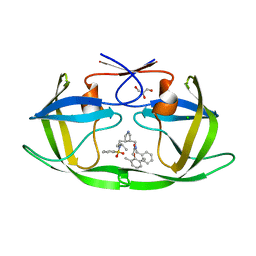 | | HIV-1 Protease in complex with pyrrolidinmethanamine | | Descriptor: | CHLORIDE ION, GLYCEROL, N-BENZYL-2-(2,6-DIMETHYLPHENOXY)-N-[((3R,4S)-4-{[ISOBUTYL(PHENYLSULFONYL)AMINO]METHYL}PYRROLIDIN-3-YL)METHYL]ACETAMIDE, ... | | Authors: | Boettcher, J, Specker, E, Heine, A, Klebe, G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Old Target Revisited: Two New Privileged Skeletons and an Unexpected Binding Mode For HIV-Protease Inhibitors
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2Y24
 
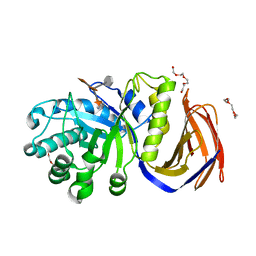 | | STRUCTURAL BASIS FOR SUBSTRATE RECOGNITION BY ERWINIA CHRYSANTHEMI GH5 GLUCURONOXYLANASE | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, IMIDAZOLE, TETRAETHYLENE GLYCOL, ... | | Authors: | Urbanikova, L, Vrsanska, M, Krogh, K.B, Hoff, T, Biely, P. | | Deposit date: | 2010-12-13 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Basis for Substrate Recognition by Erwinia Chrysanthemi Gh30 Glucuronoxylanase.
FEBS J., 278, 2011
|
|
3TEI
 
 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Remenyi, A. | | Deposit date: | 2011-08-15 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove.
Sci.Signal., 5, 2012
|
|
1IQE
 
 | | Human coagulation factor Xa in complex with M55590 | | Descriptor: | 4-[(2R)-3-[[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]AMINO]-1-OXO-2-[[[1-(4-PYRIDINYL)-4-PIPERIDINYL]METHYL]AMINO]PROPYL]-THIOMORPHOLINE-1,1-DIOXIDE, CALCIUM ION, coagulation Factor Xa | | Authors: | Shiromizu, I, Matsusue, T. | | Deposit date: | 2001-07-23 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Factor Xa Specific Inhibitor that Induces the Novel Binding Model in Complex with Human Fxa
To be Published
|
|
7LI5
 
 | | Crystal Structure Analysis of human TEAD1 | | Descriptor: | 1-[(3R,4R)-3-[4-(pyridin-3-yl)-1H-1,2,3-triazol-1-yl]-4-{[4-(trifluoromethyl)phenyl]methoxy}pyrrolidin-1-yl]prop-2-en-1-one, SULFATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure Analysis of human TEAD1
To Be Published
|
|
