6DM1
 
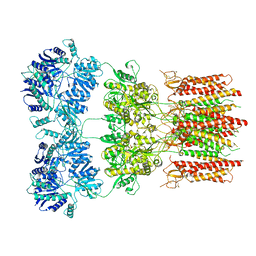 | | Open state GluA2 in complex with STZ and blocked by NASPM, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
4V3D
 
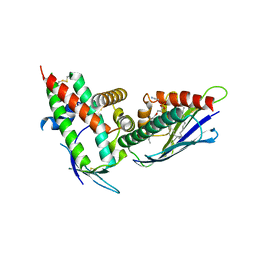 | | The CIDRa domain from HB3var03 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
6DD5
 
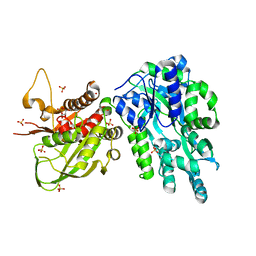 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
6DU2
 
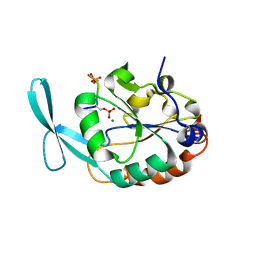 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | MAGNESIUM ION, REST-pS861/4, carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
4WCT
 
 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2014-09-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
7BC8
 
 | | Notum Fragment 658 | | Descriptor: | 1,2-ETHANEDIOL, 2-(benzyloxy)benzohydrazide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-18 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCL
 
 | | Notum Fragment 792 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BDA
 
 | | Notum Fragment 900 | | Descriptor: | (benzyloxy)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCH
 
 | | Notum Fragment 772 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-Benzyl-2-methoxyacetamide, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BD5
 
 | | Notum Fragment 830 | | Descriptor: | 2-(4-ethoxyphenyl)ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7MRW
 
 | | Native RhopH complex of the malaria parasite Plasmodium falciparum | | Descriptor: | Cytoadherence linked asexual protein 3.1, High molecular weight rhoptry protein 2, High molecular weight rhoptry protein 3 | | Authors: | Ho, C.M, Jih, J, Lai, M, Li, X.R, Goldberg, D.E, Beck, J.R, Zhou, Z.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Native structure of the RhopH complex, a key determinant of malaria parasite nutrient acquisition.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9CGT
 
 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOPENTAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-1-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
4UYW
 
 | |
7B9D
 
 | | Notum Fragment 290 | | Descriptor: | 2-(4-acetamidophenoxy)ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BAP
 
 | | Notum Fragment 648 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BDC
 
 | | Notum Fragment 923 | | Descriptor: | 1-methyl-N-[(2-methylphenyl)methyl]-1H-tetrazol-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCK
 
 | | Notum Fragment 791 | | Descriptor: | (2S)-N-(4-Methoxybenzyl)tetrahydrofuran-2-carboxamide, Palmitoleoyl-protein carboxylesterase NOTUM, SULFATE ION | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BD9
 
 | | Notum Fragment 886 | | Descriptor: | (4-chloranyl-2-methyl-pyrazol-3-yl)-piperidin-1-yl-methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
4W97
 
 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4WCX
 
 | | Crystal structure of HydG: A maturase of the [FeFe]-hydrogenase | | Descriptor: | ALANINE, Biotin and thiamin synthesis associated, FE (III) ION, ... | | Authors: | Dinis, P.C, Harmer, J.E, Driesener, R.C, Roach, P.L. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ASW
 
 | | Structure of the complex between the N-terminal dimerisation domain of Sgt2 and the UBL domain of Get5 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2, UBIQUITIN-LIKE PROTEIN MDY2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ASV
 
 | | Solution structure of the N-terminal dimerisation domain of Sgt2 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ASH
 
 | |
4AS0
 
 | | Cyclometalated Phthalimides as Protein Kinase Inhibitors | | Descriptor: | PHTALIMIDE-RUTHENIUM COMPLEX, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Blanck, S, Geisselbrecht, Y, Middel, S, Mietke, T, Harms, K, Essen, L.-O, Meggers, E. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Cyclometalated Phthalimides: Design, Synthesis and Kinase Inhibition.
Dalton Trans, 41, 2012
|
|
7ME5
 
 | | Structure of the extracellular WNT-binding module in Drl-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tyrosine-protein kinase transmembrane receptor DRL-2 | | Authors: | Shi, F, Mendrola, J.M, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
