3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
4FSE
 
 | |
3T2T
 
 | |
2PFW
 
 | |
1IWP
 
 | | Glycerol Dehydratase-cyanocobalamin Complex of Klebsiella pneumoniae | | Descriptor: | COBALAMIN, Glycerol Dehydratase Alpha subunit, Glycerol Dehydratase Beta subunit, ... | | Authors: | Yamanishi, M, Yunoki, M, Tobimatsu, T, Toraya, T. | | Deposit date: | 2002-05-28 | | Release date: | 2002-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol.
Eur.J.Biochem., 269, 2002
|
|
2PR7
 
 | |
3O3E
 
 | | Human Class I MHC HLA-A2 in complex with the Peptidomimetic ELA-2.1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Borbulevych, O.Y, Baker, B.M. | | Deposit date: | 2010-07-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of HLA-A*0201 complexed with Melan-A/MART-1(26(27L)-35) peptidomimetics reveal conformational heterogeneity and highlight degeneracy of T cell recognition.
J.Med.Chem., 53, 2010
|
|
2G97
 
 | |
3UIX
 
 | |
4FSL
 
 | |
1SLM
 
 | |
2QPX
 
 | |
1JH1
 
 | | Crystal Structure of MMP-8 complexed with a 6H-1,3,4-thiadiazine derived inhibitor | | Descriptor: | BUT-3-ENYL-[5-(4-CHLORO-PHENYL)-3,6-DIHYDRO-[1,3,4]THIADIAZIN-2-YLIDENE]-AMINE, CALCIUM ION, Matrix Metalloproteinase 8, ... | | Authors: | Schroder, J, Henke, A, Wenzel, H, Brandstetter, H, Stammler, H.G, Stammler, A, Pfeiffer, W.D, Tschesche, H. | | Deposit date: | 2001-06-27 | | Release date: | 2001-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based design and synthesis of potent matrix metalloproteinase inhibitors derived from a 6H-1,3,4-thiadiazine scaffold.
J.Med.Chem., 44, 2001
|
|
2QTD
 
 | |
4A9E
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 3-methyl-1,2,3,4- tetrahydroquinazolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, BROMODOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 1: Inhibitor Binding Modes and Implications for Lead Discovery.
J.Med.Chem., 55, 2012
|
|
4A9L
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 1,3-DIMETHYL-6-(MORPHOLINE- 4-SULFONYL)-1,2,3,4-TETRAHYDROQUINAZOLIN-2-ONE | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-6-(morpholin-4-ylsulfonyl)-3,4-dihydroquinazolin-2(1H)-one, BROMODOMAIN-CONTAINING PROTEIN 4, ... | | Authors: | Chung, C.W, Bamborough, P. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Discovery of Bromodomain Inhibitors Part 1: Inhibitor Binding Modes and Implications for Lead Discovery.
J.Med.Chem., 55, 2012
|
|
3ZWA
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-HEXANOYL-COA | | Descriptor: | (S)-3-Hydroxyhexanoyl-CoA, GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3G8Y
 
 | |
1PG3
 
 | | Acetyl CoA Synthetase, Acetylated on Lys609 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, COENZYME A, ... | | Authors: | Gulick, A.M, Starai, V.J, Horswill, A.R, Homick, K.M, Escalante-Semerena, J.C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.75 A Crystal Structure of Acetyl-CoA Synthetase Bound
to Adenosine-5'-propylphosphate and Coenzyme A
Biochemistry, 42, 2003
|
|
1PG4
 
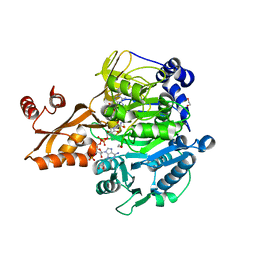 | | Acetyl CoA Synthetase, Salmonella enterica | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, CHLORIDE ION, ... | | Authors: | Gulick, A.M, Starai, V.J, Horswill, A.R, Homick, K.M, Escalante-Semerena, J.C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.75 A Crystal Structure of Acetyl-CoA Synthetase Bound
to Adenosine-5'-propylphosphate and Coenzyme A
Biochemistry, 42, 2003
|
|
1YHS
 
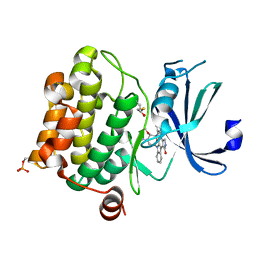 | | Crystal structure of Pim-1 bound to staurosporine | | Descriptor: | Proto-oncogene serine/threonine-protein kinase Pim-1, STAUROSPORINE | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1OIQ
 
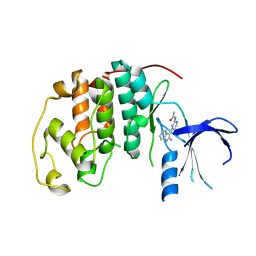 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-[4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)-2-PYRIMIDINYL]ACETAMIDE | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
3VIG
 
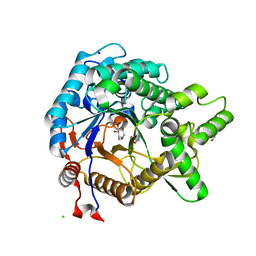 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with 1-deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-glucosidase, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2C53
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | 2'-DEOXYURIDINE, GLYCEROL, URACIL DNA GLYCOSYLASE | | Authors: | Krusong, K, Carpenter, E.P, Bellmy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2SNS
 
 | | STAPHYLOCOCCAL NUCLEASE. PROPOSED MECHANISM OF ACTION BASED ON STRUCTURE OF ENZYME-THYMIDINE 3(PRIME),5(PRIME)-BIPHOSPHATE-CALCIUM ION COMPLEX AT 1.5-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, THERMONUCLEASE PRECURSOR, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Legg, M.J, Cotton, F.A, Hazen Jr, E.E. | | Deposit date: | 1982-05-14 | | Release date: | 1982-07-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | STAPHYLOCOCCAL NUCLEASE. PROPOSED MECHANISM OF ACTION BASED ON STRUCTURE OF ENZYME-THYMIDINE 3(PRIME),5(PRIME)-BIPHOSPHATE-CALCIUM ION COMPLEX AT 1.5-ANGSTROMS RESOLUTION
Thesis, Texas Agricultural and Mechanical University, 1977
|
|
