8AJZ
 
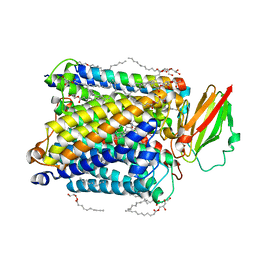 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase at 2 milliseconds after irradiation by a 532 nm laser | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Bosman, R, Dahl, P, Nango, E, Tanaka, R, Zoric, D, Svensson, E, Nakane, T, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
5G0B
 
 | | An unusual natural product primary sulfonamide: synthesis, carbonic anhydrase inhibition and protein x-ray structure of Psammaplin C | | Descriptor: | CARBONIC ANHYDRASE 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mujumdar, P, Supuran, C.T, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2016-03-17 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition and Protein X-Ray Structures of Psammaplin C.
J.Med.Chem., 59, 2016
|
|
5GSK
 
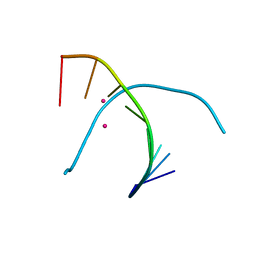 | | Crystal structure of duplex DNA3 in complex with Hg(II) and Sr(II) | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION, STRONTIUM ION | | Authors: | Liu, H.H, Wang, R, Yao, Q.Q, Cheng, Y.Q, Yang, C, Luo, Q, Wu, B.X, Li, J.X, Ma, J.B, Sheng, J, Gan, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5PZP
 
 | |
5WSQ
 
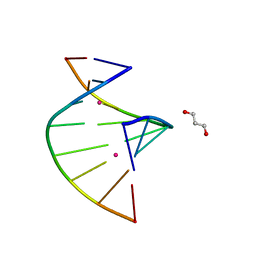 | | Crystal structure of C-Hg-T pair containing DNA duplex | | Descriptor: | 1,3-PROPANDIOL, DNA (5'-D(*GP*CP*CP*CP*GP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5E4F
 
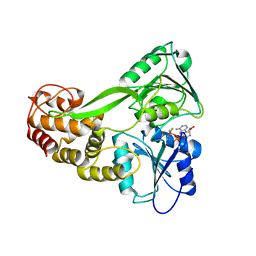 | |
5UOY
 
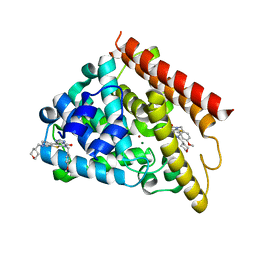 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 16j (6-(4-Methoxybenzyl)-9-((tetrahydro-2H-pyran-4-yl)methyl)-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-methoxyphenyl)methyl]-9-[(oxan-4-yl)methyl]-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
7KHP
 
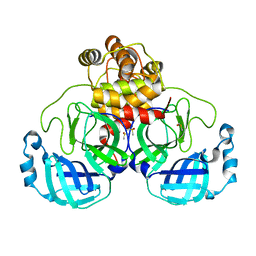 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
3G01
 
 | |
1TRL
 
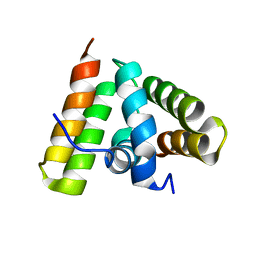 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
1I2T
 
 | |
1BZT
 
 | |
5BND
 
 | | Crystal structure of the C-terminal domain of TagH | | Descriptor: | ABC transporter, ATP-binding protein | | Authors: | Chen, S.C, Chen, Y. | | Deposit date: | 2015-05-26 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SH3-Like Motif-Containing C-terminal Domain of Staphylococcal Teichoic Acid Transporter Suggests Possible Function.
Proteins, 2016
|
|
4MK7
 
 | |
6PCM
 
 | |
4MK8
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 4 (N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
2VH4
 
 | | Structure of a loop C-sheet serpin polymer | | Descriptor: | TENGPIN | | Authors: | Zhang, Q, Law, R.H.P, Bottomley, S.P, Whisstock, J.C, Buckle, A.M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Structural Basis for Loop C-Sheet Polymerization in Serpins.
J.Mol.Biol., 376, 2008
|
|
4MKA
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 13 (N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{3-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1-oxo-1H-isochromen-7-yl}methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
6TDB
 
 | | Neuropilin2-b1 domain in a complex with the C-terminal VEGFB167 peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, C-terminal VEGFB167 peptide, ... | | Authors: | Eldrid, C, Yu, L, Yelland, T, Fotinou, C, Djordjevic, S. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NRP2-b1 domain in a complex with the C-terminal VEGFB167 peptide
To Be Published
|
|
4PTD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT D274N | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
2WD1
 
 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1LKV
 
 | |
4AEX
 
 | |
6AU4
 
 | | Crystal structure of the major quadruplex formed in the human c-MYC promoter | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), POTASSIUM ION | | Authors: | Stump, S, Mou, T.C, Sprang, S.R, Natale, N.R, Beall, H.D. | | Deposit date: | 2017-08-30 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene.
PLoS ONE, 13, 2018
|
|
7LRG
 
 | |
