3SRY
 
 | |
3SS0
 
 | |
3SG7
 
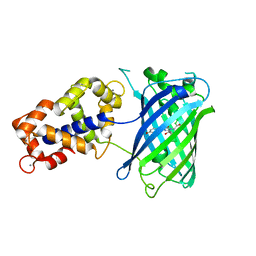 | | Crystal Structure of GCaMP3-KF(linker 1) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG5
 
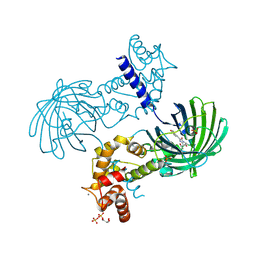 | | Crystal Structure of Dimeric GCaMP3-D380Y, QP(linker 1), LP(linker 2) | | Descriptor: | CALCIUM ION, GLYCEROL, Myosin light chain kinase, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG2
 
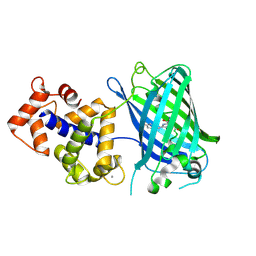 | | Crystal Structure of GCaMP2-T116V,D381Y | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG6
 
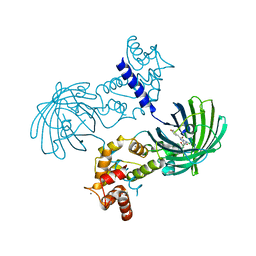 | | Crystal Structure of Dimeric GCaMP2-LIA(linker 1) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG3
 
 | | Crystal Structure of GCaMP3-D380Y | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG4
 
 | | Crystal Structure of GCaMP3-D380Y, LP(linker 2) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3S05
 
 | |
3RWT
 
 | |
3RWA
 
 | |
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
2YE1
 
 | | X-ray structure of the cyan fluorescent proteinmTurquoise-GL (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of the Cyan Fluorescent Protein Mturquoise-Gl
To be Published
|
|
2YDZ
 
 | | X-ray structure of the cyan fluorescent protein SCFP3A (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
2Y0G
 
 | |
3PJB
 
 | |
3PJ5
 
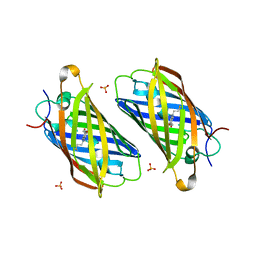 | |
3PJ7
 
 | |
3PIB
 
 | |
3P8U
 
 | | Crystal structure of mEosFP in its green state | | Descriptor: | Green to red photoconvertible GPF-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2010-10-15 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational design of photoconvertible and biphotochromic fluorescent proteins for advanced microscopy applications.
Chem.Biol., 18, 2011
|
|
3OSR
 
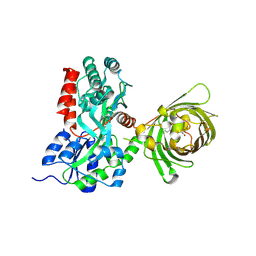 | | Maltose-bound maltose sensor engineered by insertion of circularly permuted green fluorescent protein into E. coli maltose binding protein at position 311 | | Descriptor: | Maltose-binding periplasmic protein,Green fluorescent protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Echevarria, I.M, Marvin, J.S, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2010-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A genetically encoded, high-signal-to-noise maltose sensor.
Proteins, 79, 2011
|
|
3OSQ
 
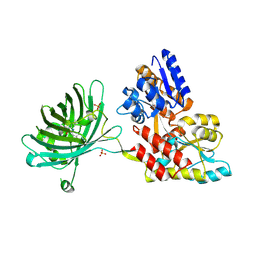 | | Maltose-bound maltose sensor engineered by insertion of circularly permuted green fluorescent protein into E. coli maltose binding protein at position 175 | | Descriptor: | Maltose-binding periplasmic protein,Green fluorescent protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Echevarria, I.M, Marvin, J.S, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2010-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A genetically encoded, high-signal-to-noise maltose sensor.
Proteins, 79, 2011
|
|
3OGO
 
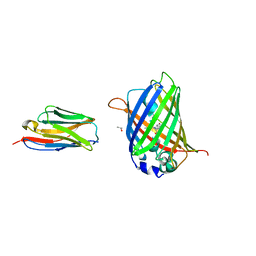 | | Structure of the GFP:GFP-nanobody complex at 2.8 A resolution in spacegroup P21212 | | Descriptor: | GFP-nanobody, Green fluorescent protein, ISOPROPYL ALCOHOL | | Authors: | Kubala, M.H, Kovtun, O, Alexandrov, K, Collins, B.M. | | Deposit date: | 2010-08-17 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and thermodynamic analysis of the GFP:GFP-nanobody complex.
Protein Sci., 19, 2010
|
|
3O78
 
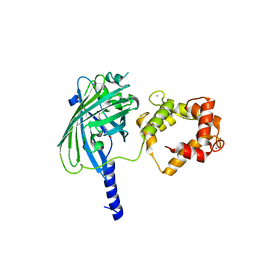 | | The structure of Ca2+ Sensor (Case-12) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, smooth muscle,Green fluorescent protein,Green fluorescent protein,Calmodulin-1 | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
3O77
 
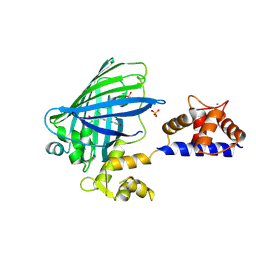 | | The structure of Ca2+ Sensor (Case-16) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Myosin light chain kinase, ... | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
