3SG7
 
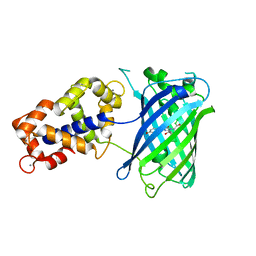 | | Crystal Structure of GCaMP3-KF(linker 1) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG5
 
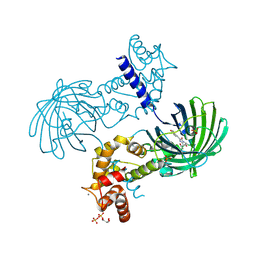 | | Crystal Structure of Dimeric GCaMP3-D380Y, QP(linker 1), LP(linker 2) | | Descriptor: | CALCIUM ION, GLYCEROL, Myosin light chain kinase, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
4TZA
 
 | |
5FVG
 
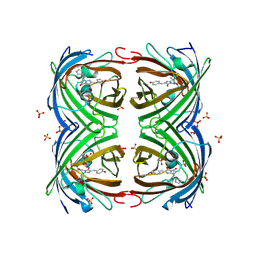 | | Structure of IrisFP at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett, 7, 2016
|
|
4KKM
 
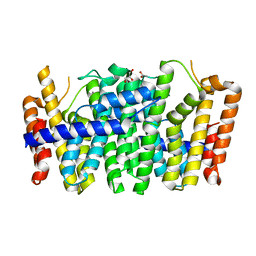 | | Crystal structure of a FPP/GFPP synthase (Target EFI-501952) from Zymomonas mobilis, apo structure | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a FPP/GFPP synthase (Target EFI-501952) from Zymomonas mobilis, apo structure
To be Published
|
|
1EMA
 
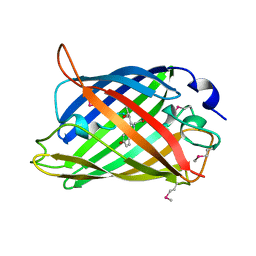 | |
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
4QEC
 
 | | ElxO with NADP Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
4ZF3
 
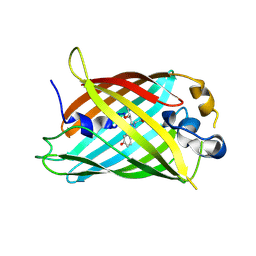 | |
5UKD
 
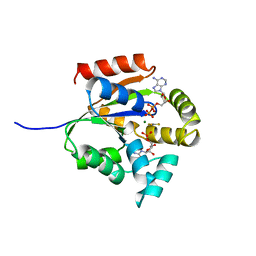 | |
5AB6
 
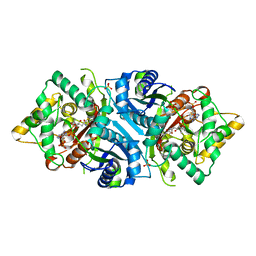 | |
3GJ2
 
 | | Photoactivated state of PA-GFP | | Descriptor: | CHLORIDE ION, Green fluorescent protein | | Authors: | Henderson, J.N, Gepshtein, R, Heenan, J.R, Kallio, K, Huppert, D, Remington, S.J. | | Deposit date: | 2009-03-07 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the photoactivatable green fluorescent protein.
J.Am.Chem.Soc., 131, 2009
|
|
1GFL
 
 | |
1UKZ
 
 | |
5EJE
 
 | | Crystal structure of E. coli Adenylate kinase G56C/T163C double mutant in complex with Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, COBALT (II) ION | | Authors: | Sauer, U.H, Kovermann, M, Grundstrom, C, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-01 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for ligand binding to an enzyme by a conformational selection pathway.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2GX0
 
 | |
8U20
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 5 | | Descriptor: | Green Fluorescent Protein Variant #5, ccGFP 5 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
5LOD
 
 | |
7LYI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
1ZGQ
 
 | |
4H47
 
 | | 1.9 angstrom CyPet structure at pH5.2 | | Descriptor: | ACETATE ION, Green fluorescent protein, SULFATE ION | | Authors: | Hu, X.-J, Liu, R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights of the fluorescent states of CyPet
To be Published
|
|
2VMA
 
 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F, IODIDE ION | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
4Q9X
 
 | | mTFP* PdCl2 soak | | Descriptor: | CARBONATE ION, CHLORIDE ION, GFP-like fluorescent chromoprotein cFP484, ... | | Authors: | Fischer, J, Quitterer, F, Groll, M, Eppinger, J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | mTFP*: An imperishable and versatile protein host for anchoring diverse ligands and organocatalysts
To be Published
|
|
2OKW
 
 | |
