1WY3
 
 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH7.0 | | Descriptor: | Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8CNF
 
 | |
6Y0Y
 
 | | Sarcin Ricin Loop, mutant C2666U | | Descriptor: | MAGNESIUM ION, RNA (27-MER), SODIUM ION | | Authors: | Ennifar, E, Westhof, E. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Sarcin Ricin Loop, mutant C2666U
To Be Published
|
|
6FO5
 
 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR #17 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[4-[[7-ethyl-2,6-bis(oxidanylidene)purin-3-yl]methyl]phenyl]methyl]-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepine-7-sulfonamide | | Authors: | Raux, B, Betzi, S. | | Deposit date: | 2018-02-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
6WEY
 
 | |
4QIO
 
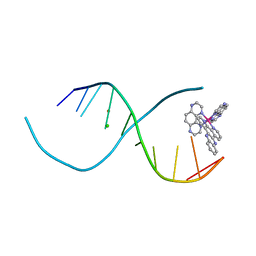 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(TCGGCGCCIA) at high resolution | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*IP*A)-3', BARIUM ION, CHLORIDE ION, ... | | Authors: | Gurung, S.P, Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-06-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Inosine Can Increase DNA's Susceptibility to Photo-oxidation by a Ru II Complex due to Structural Change in the Minor Groove.
Chemistry, 23, 2017
|
|
4F19
 
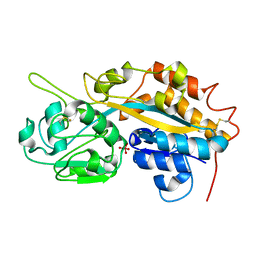 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 4.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
6TJ9
 
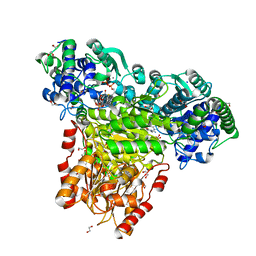 | | Escherichia coli transketolase in complex with cofactor analog 2'-methoxythiamine and substrate xylulose 5-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-azanyl-2-methoxy-pyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 5-O-phosphono-D-xylulose, ... | | Authors: | Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2019-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for antibiotic action of the B 1 antivitamin 2'-methoxy-thiamine.
Nat.Chem.Biol., 16, 2020
|
|
5E9N
 
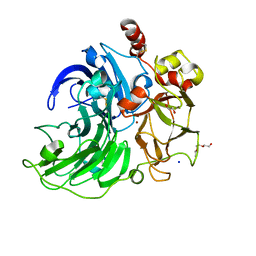 | | Steccherinum murashkinskyi laccase at 0.95 resolution | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Glazunova, O.A, Fedorova, T.V, Koroleva, O.V. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi.
Int. J. Biol. Macromol., 118, 2018
|
|
4WKA
 
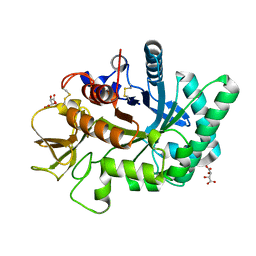 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8SY4
 
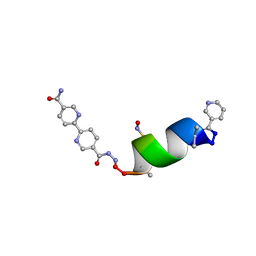 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-SER-LEU-ALA-SNC-AIB-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-05-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
1NKI
 
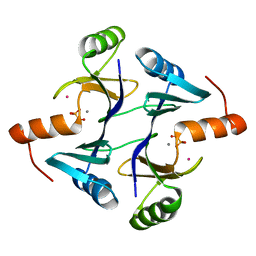 | | CRYSTAL STRUCTURE OF THE FOSFOMYCIN RESISTANCE PROTEIN A (FOSA) CONTAINING BOUND PHOSPHONOFORMATE | | Descriptor: | MANGANESE (II) ION, PHOSPHONOFORMIC ACID, POTASSIUM ION, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2003-01-03 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA.
Biochemistry, 43, 2004
|
|
7FDX
 
 | | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe 8-Anilino-1-naphthalenesulfonic acid (ANS) | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe 8-Anilino-1-naphthalenesulfonic acid (ANS)
To Be Published
|
|
7FEU
 
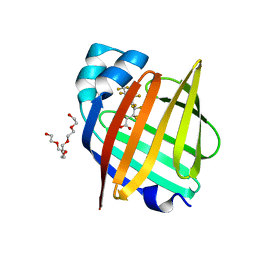 | | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorononanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Hara, T, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluorononanoic acid
To Be Published
|
|
6TXP
 
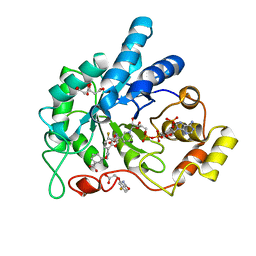 | | Human Aldose Reductase Mutant L300A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid) | | Descriptor: | (2-carbamoyl-5-fluorophenoxy)acetic acid, 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldo-keto reductase family 1 member B1, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Human Aldose Reductase Mutant L300A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid)
To Be Published
|
|
1OB4
 
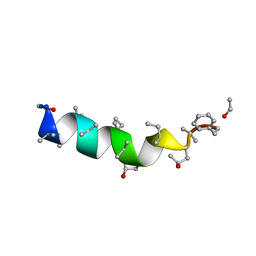 | | Cephaibol A | | Descriptor: | CEPHAIBOL A, ETHANOL | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
352D
 
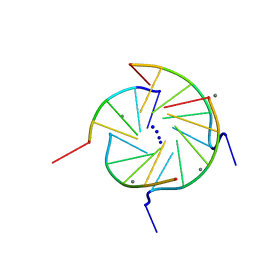 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
7LTV
 
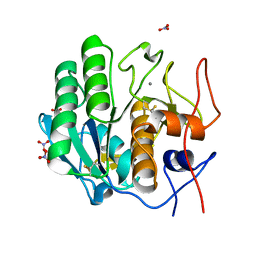 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 3 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTB
 
 | | Crystal Structure of Keratinicyclin B | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, 3-ammonio-2,3,6-trideoxy-alpha-L-arabino-hexopyranose-(1-2)-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-19 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Potent and specific antibiotic combination therapy against Clostridioides difficile.
Nat.Chem.Biol., 20, 2024
|
|
5NMN
 
 | |
6TWT
 
 | | Crystal structure of N-terminally truncated NDM-1 metallo-beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Imiolczyk, B, Czyrko-Horczak, J, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
7QQC
 
 | | Structure of CTX-M-15 K73A mutant | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
3DK9
 
 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, SULFATE ION | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
1DPN
 
 | | B-DODECAMER CGCGAA(TAF)TCGCG WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Teplova, M, Minasov, G, Joachimiak, A, Sanishvili, R, Weeks, C.M, Miller, R, Maier, M.A, An, H, Dan Cook, P, Manoharan, M. | | Deposit date: | 1999-12-27 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution.
Biopolymers, 48, 1998
|
|
4LZT
 
 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
