4JFG
 
 | | Crystal structure of sfGFP-66-HqAla | | Descriptor: | CESIUM ION, Green fluorescent protein, quinolin-8-ol | | Authors: | Wang, J, Liu, X, Li, J, Zhang, W, Hu, M, Zhou, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Significant expansion of the fluorescent protein chromophore through the genetic incorporation of a metal-chelating unnatural amino acid.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4JGE
 
 | |
4JKY
 
 | |
4JL5
 
 | |
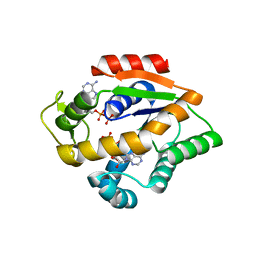
4JL6
 
 | |
4JL8
 
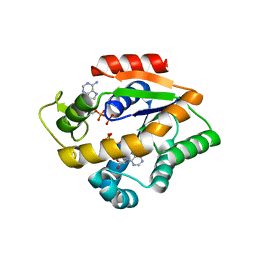 | |
4JLA
 
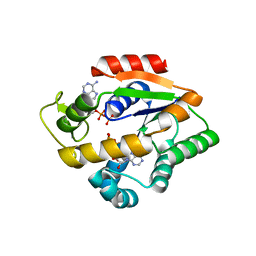 | |
4JLB
 
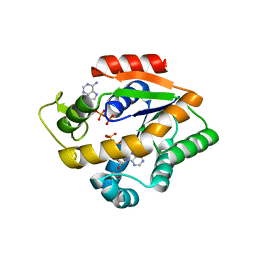 | |
4JLD
 
 | |
4JLO
 
 | |
4JLP
 
 | |
4JQ7
 
 | | Crystal structure of EGFR kinase domain in complex with compound 2a | | Descriptor: | (2S)-2-[(5,6-diphenylfuro[2,3-d]pyrimidin-4-yl)amino]-2-phenylethanol, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JQ8
 
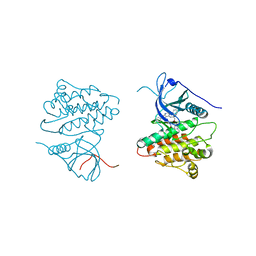 | | Crystal structure of EGFR kinase domain in complex with compound 4b | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]-N~3~,N~3~-dimethyl-beta-alaninamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JR3
 
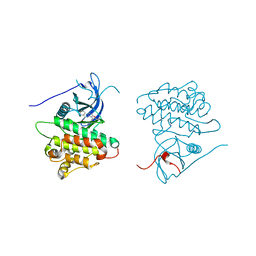 | | Crystal structure of EGFR kinase domain in complex with compound 3g | | Descriptor: | Epidermal growth factor receptor, N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]acetamide | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JRB
 
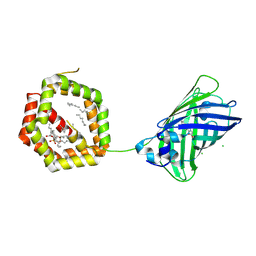 | | Structure of Cockroach Allergen Bla g 1 Tandem Repeat as a EGFP fusion | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CHLORIDE ION, DODECANE, ... | | Authors: | Mueller, G.A, Pedersen, L.C, Lih, F.B, Glesner, J, Moon, A.F, Chapman, M.D, Tomer, K, London, R.E. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment.
J.Allergy Clin.Immunol., 132, 2013
|
|
4JRV
 
 | | Crystal structure of EGFR kinase domain in complex with compound 4c | | Descriptor: | 4-(dimethylamino)-N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]butanamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4JZK
 
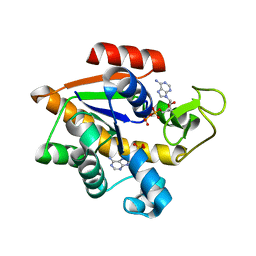 | |
4K46
 
 | | Crystal Structure of Adenylate Kinase from Photobacterium profundum | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, ... | | Authors: | Cho, Y.-J, Kerns, S.J, Kern, D. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Alike but Different: Adenylate kinases from E. Coli, Aquifex, and P. profundum
To be Published
|
|
4KA9
 
 | |
4KAG
 
 | | Crystal structure analysis of a single amino acid deletion mutation in EGFP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Arpino, J.A.J, Rizkallah, P.J. | | Deposit date: | 2013-04-22 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and dynamic changes associated with beneficial engineered single-amino-acid deletion mutations in enhanced green fluorescent protein.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KEX
 
 | |
4KF4
 
 | | Crystal Structure of sfCherry | | Descriptor: | fluorescent protein sfCherry | | Authors: | Nguyen, H.B, Hung, L.-W, Yeates, T.O, Waldo, G.S, Terwilliger, T.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Split green fluorescent protein as a modular binding partner for protein crystallization.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KF5
 
 | | Crystal Structure of Split GFP complexed with engineered sfCherry with an insertion of GFP fragment | | Descriptor: | fluorescent protein GFP1-9, fluorescent protein sfCherry+GFP10-11 | | Authors: | Nguyen, H.B, Hung, L.-W, Yeates, T.O, Waldo, G.S, Terwilliger, T.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Split green fluorescent protein as a modular binding partner for protein crystallization.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KGE
 
 | | Crystal structure of near-infrared fluorescent protein with an extended stokes shift, pH 4.5 | | Descriptor: | CHLORIDE ION, TagRFP675, red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant.
Sci Rep, 3, 2013
|
|
4KGF
 
 | | Crystal structure of near-infrared fluorescent protein with an extended stokes shift, ph 8.0 | | Descriptor: | CHLORIDE ION, TagRFP675, red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant.
Sci Rep, 3, 2013
|
|
