1BQ3
 
 | |
2GFJ
 
 | | Crystal structure of the zinc-beta-lactamase L1 from stenotrophomonas maltophilia (inhibitor 1) | | Descriptor: | 1,3-DIPHENYL-1H-PYRAZOLE-4,5-DICARBOXYLIC ACID, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
2H5K
 
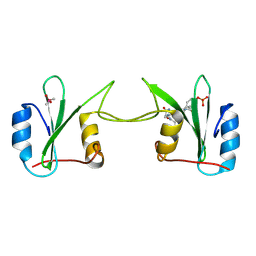 | | Crystal Structure of Complex Between the Domain-Swapped Dimeric Grb2 SH2 Domain and Shc-Derived Ligand, Ac-NH-pTyr-Val-Asn-NH2 | | Descriptor: | CACODYLATE ION, Growth factor receptor-bound protein 2, Shc-Derived Ligand | | Authors: | Benfield, A.P, Whiddon, B.B, Martin, S.F. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural and energetic aspects of Grb2-SH2 domain-swapping.
Arch.Biochem.Biophys., 462, 2007
|
|
1BQ4
 
 | |
3D1K
 
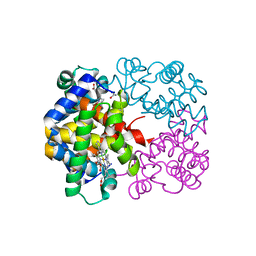 | | R/T intermediate quaternary structure of an antarctic fish hemoglobin in an alpha(CO)-beta(pentacoordinate) state | | Descriptor: | ACETYL GROUP, CARBON MONOXIDE, Hemoglobin subunit alpha-1, ... | | Authors: | Vitagliano, L, Vergara, A, Bonomi, G, Merlino, A, Mazzarella, L. | | Deposit date: | 2008-05-06 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Spectroscopic and crystallographic characterization of a tetrameric hemoglobin oxidation reveals structural features of the functional intermediate relaxed/tense state.
J.Am.Chem.Soc., 130, 2008
|
|
2H6A
 
 | | Crystal structure of the zinc-beta-lactamase L1 from Stenotrophomonas maltophilia (mono zinc form) | | Descriptor: | Metallo-beta-lactamase L1, SULFATE ION, ZINC ION | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-05-31 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
7VQS
 
 | | Crystal structure of LSD1 in complex with compound 4 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQU
 
 | | Crystal structure of LSD1 in complex with compound S1427 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
1X8G
 
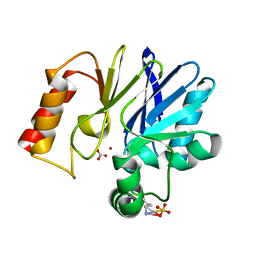 | |
8A31
 
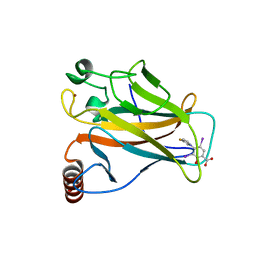 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC694 | | Descriptor: | 4-(3-fluoranylpyrrol-1-yl)-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8A32
 
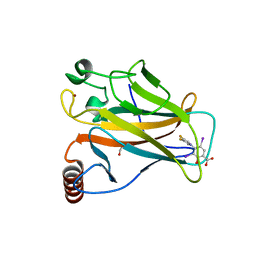 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC769 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3,4-bis(fluoranyl)pyrrol-1-yl]-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
1BTT
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BTR
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1993-05-25 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1BOC
 
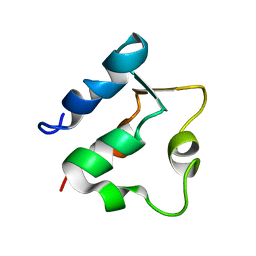 | | THE SOLUTION STRUCTURES OF MUTANT CALBINDIN D9K'S, AS DETERMINED BY NMR, SHOW THAT THE CALCIUM BINDING SITE CAN ADOPT DIFFERENT FOLDS | | Descriptor: | CALBINDIN D9K | | Authors: | Johansson, C, Ullner, M, Drakenberg, T. | | Deposit date: | 1993-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of mutant calbindin D9k's, as determined by NMR, show that the calcium-binding site can adopt different folds.
Biochemistry, 32, 1993
|
|
1BXR
 
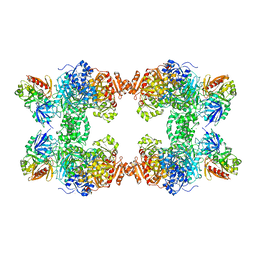 | | STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE COMPLEXED WITH THE ATP ANALOG AMPPNP | | Descriptor: | CARBAMOYL-PHOSPHATE SYNTHASE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Wesenberg, G, Raushel, F.M, Holden, H.M. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding.
Biochemistry, 38, 1999
|
|
1BVD
 
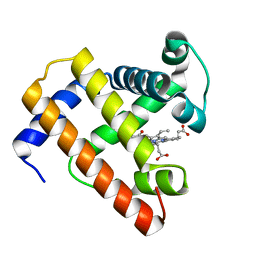 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM B) AT 98 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
1BVC
 
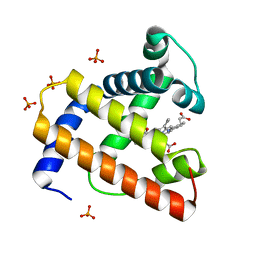 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM D) AT 118 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA, PHOSPHATE ION | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
1BOD
 
 | | THE SOLUTION STRUCTURES OF MUTANT CALBINDIN D9K'S, AS DETERMINED BY NMR, SHOW THAT THE CALCIUM BINDING SITE CAN ADOPT DIFFERENT FOLDS | | Descriptor: | CALBINDIN D9K | | Authors: | Johansson, C, Ullner, M, Drakenberg, T. | | Deposit date: | 1993-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The solution structures of mutant calbindin D9k's, as determined by NMR, show that the calcium-binding site can adopt different folds.
Biochemistry, 32, 1993
|
|
2JHF
 
 | | Structural evidence for a ligand coordination switch in liver alcohol dehydrogenase | | Descriptor: | ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meijers, R, Adolph, H.W, Dauter, Z, Wilson, K.S, Lamzin, V.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Evidence for a Ligand Coordination Switch in Liver Alcohol Dehydrogenase
Biochemistry, 46, 2007
|
|
1CK7
 
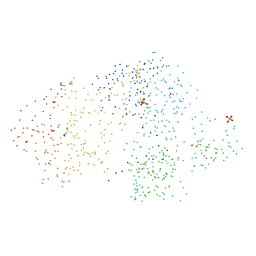 | | GELATINASE A (FULL-LENGTH) | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (GELATINASE A), ... | | Authors: | Morgunova, E, Tuuttila, A, Bergmann, U, Isupov, M, Lindqvist, Y, Schneider, G, Tryggvason, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed.
Science, 284, 1999
|
|
7VJT
 
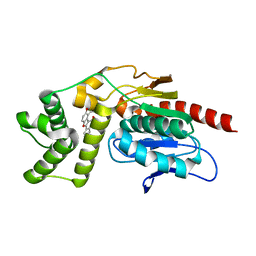 | | Crystal Structure of Mtb Pks13-TE in complex with inhibitor coumestan derivative 8 | | Descriptor: | 3,8-bis(oxidanyl)-7-(piperidin-1-ylmethyl)-[1]benzofuro[3,2-c]chromen-6-one, Polyketide synthase Pks13 (Termination polyketide synthase) | | Authors: | Zhang, W, Wang, S.S, Yu, L.F. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of Coumestan Derivatives as Polyketide Synthase 13-Thioesterase(Pks13-TE) Inhibitors with Improved hERG Profiles for Mycobacterium tuberculosis Treatment.
J.Med.Chem., 65, 2022
|
|
1CR1
 
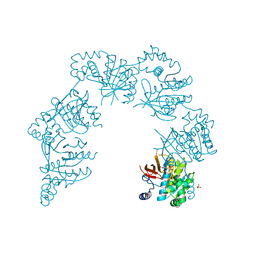 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTTP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR4
 
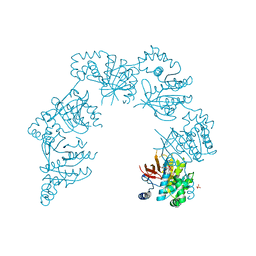 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
7OC7
 
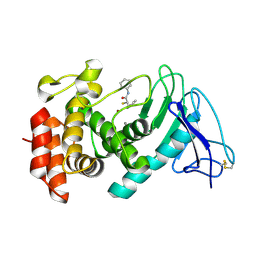 | | LasB, alpha-alkyl-N-aryl mercaptoacetamide | | Descriptor: | (2R)-N,3-diphenyl-2-sulfanyl-propanamide, CALCIUM ION, Neutral metalloproteinase, ... | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2021-04-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Inspired Fragment Merging and Growing Affords Efficacious LasB Inhibitors.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1CR2
 
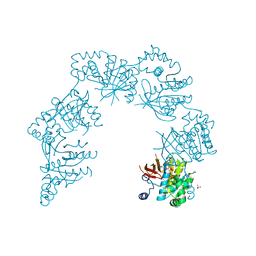 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
