3IWI
 
 | |
3IWO
 
 | |
3IXG
 
 | |
3IXH
 
 | |
3IXB
 
 | |
3IWQ
 
 | |
3IXD
 
 | |
3IIU
 
 | | Structure of the reconstituted Peridinin-Chlorophyll a-Protein (RFPCP) mutant N89L | | Descriptor: | (2S)-3-[(6-O-alpha-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-2-[(3Z,6Z,9Z,12Z,15Z)-octadeca-3,6,9,12,15-pentaenoyloxy]propyl (5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Schulte, T, Hofmann, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of a single peridinin sensing Chl-a excitation in reconstituted PCP by crystallography and spectroscopy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5UJB
 
 | | Structure of a Mcl-1 Inhibitor Binding to Site 3 of Human Serum Albumin | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, PHOSPHATE ION, Serum albumin | | Authors: | Zhao, B. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Myeloid cell leukemia-1 (Mcl-1) inhibitor bound to drug site 3 of Human Serum Albumin.
Bioorg. Med. Chem., 25, 2017
|
|
3IIS
 
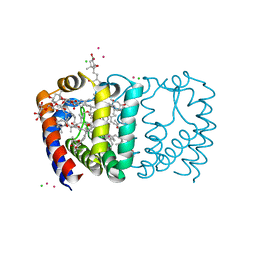 | | Structure of the reconstituted Peridinin-Chlorophyll a-Protein (RFPCP) | | Descriptor: | (2S)-3-[(6-O-alpha-D-galactopyranosyl-beta-D-galactopyranosyl)oxy]-2-[(3Z,6Z,9Z,12Z,15Z)-octadeca-3,6,9,12,15-pentaenoyloxy]propyl (5Z,8Z,11Z,14Z,17Z)-icosa-5,8,11,14,17-pentaenoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Schulte, T, Hofmann, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of a single peridinin sensing Chl-a excitation in reconstituted PCP by crystallography and spectroscopy.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1M35
 
 | | Aminopeptidase P from Escherichia coli | | Descriptor: | AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Graham, S.C, Lee, M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-06-27 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3IQW
 
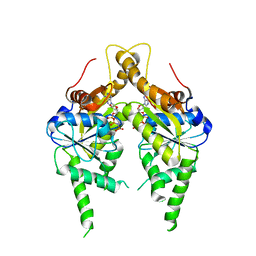 | | AMPPNP complex of C. therm. Get3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3NX3
 
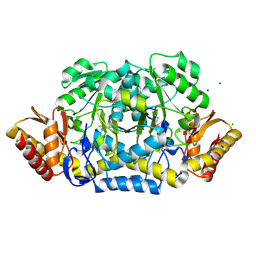 | | Crystal structure of acetylornithine aminotransferase (argD) from Campylobacter jejuni | | Descriptor: | Acetylornithine aminotransferase, MAGNESIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
8EJN
 
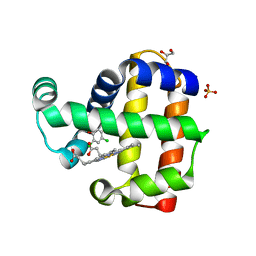 | | Structure of dehaloperoxidase A in complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, DIMETHYL SULFOXIDE, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Franzen, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Comparative study of the binding and activation of 2,4-dichlorophenol by dehaloperoxidase A and B.
J.Inorg.Biochem., 247, 2023
|
|
3P2O
 
 | | Crystal Structure of FolD Bifunctional Protein from Campylobacter jejuni | | Descriptor: | Bifunctional protein folD, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kim, Y, Zhang, R, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Crystal Structure of FolD Bifunctional Protein from
To be Published
|
|
6S6N
 
 | |
6S6M
 
 | |
3PGM
 
 | | THE STRUCTURE OF YEAST PHOSPHOGLYCERATE MUTASE AT 0.28 NM RESOLUTION | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate mutase 1, SULFATE ION | | Authors: | Campbell, J.W, Hodgson, G.I, Warwicker, J, Winn, S.I, Watson, H.C. | | Deposit date: | 1982-04-06 | | Release date: | 1982-05-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of phosphoglycerate mutase.
Philos.Trans.R.Soc.London,Ser.B, 293, 1981
|
|
8G1N
 
 | | Structure of Campylobacter concisus PglC I57M/Q175M Variant with modeled C-terminus | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MAGNESIUM ION, N,N'-diacetylbacilliosaminyl-1-phosphate transferase, ... | | Authors: | Dodge, G.J, Ray, L.C, Das, D, Imperiali, B, Allen, K.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Co-conserved sequence motifs are predictive of substrate specificity in a family of monotopic phosphoglycosyl transferases.
Protein Sci., 32, 2023
|
|
7MNH
 
 | |
1BLS
 
 | |
3RCE
 
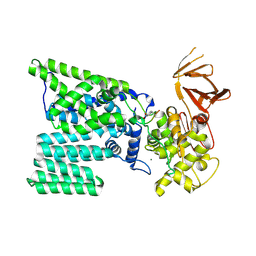 | | Bacterial oligosaccharyltransferase PglB | | Descriptor: | MAGNESIUM ION, Oligosaccharide transferase to N-glycosylate proteins, Substrate Mimic Peptide | | Authors: | Lizak, C, Gerber, S, Numao, S, Aebi, M, Locher, K.P. | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a bacterial oligosaccharyltransferase.
Nature, 474, 2011
|
|
1KFR
 
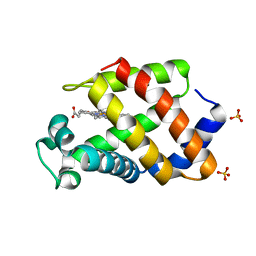 | | Structural plasticity in the eight-helix fold of a trematode hemoglobin | | Descriptor: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Milani, M, Pesce, A, Dewilde, S, Ascenzi, P, Moens, L, Bolognesi, M. | | Deposit date: | 2001-11-22 | | Release date: | 2002-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural plasticity in the eight-helix fold of a trematode haemoglobin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4HXX
 
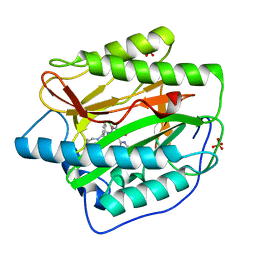 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
2Q9M
 
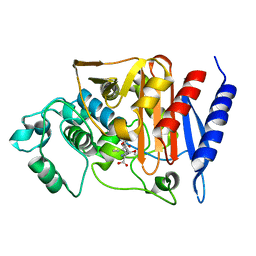 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
