3DFU
 
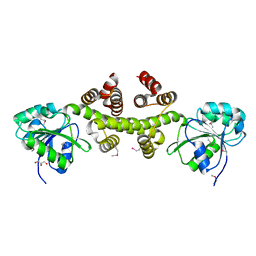 | |
4CBZ
 
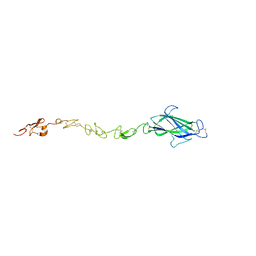 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN JAGGED-1, alpha-L-fucopyranose | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
2DAL
 
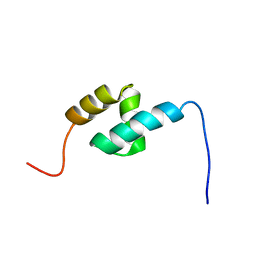 | | Solution Structure of the Novel Identified UBA-like Domain in the N-terminal of Human Fas Associated Factor 1 Protein | | Descriptor: | Protein KIAA0794 | | Authors: | Zhao, C, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Novel Identified UBA-like Domain in the N-terminal of Human Fas Associated Factor 1 Protein
To be Published
|
|
7GYF
 
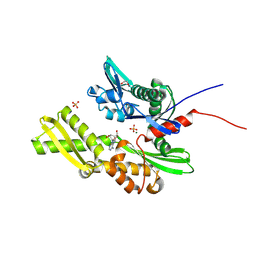 | | Crystal Structure of HSP72 in complex with ligand 10 at 13.68 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GYH
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 15.96 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
4MNF
 
 | | Crystal structure of BRAF-V600E bound to GDC0879 | | Descriptor: | 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Sudhamsu, J, Haling, J.R, Morales, T, Brandhuber, B, Hymowitz, S.G. | | Deposit date: | 2013-09-10 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the BRAF-MEK Complex Reveals a Kinase Activity Independent Role for BRAF in MAPK Signaling.
Cancer Cell, 26, 2014
|
|
3GOK
 
 | | Binding site mapping of protein ligands | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 2 | | Authors: | Scheich, C. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Binding site mapping of protein ligands
To be published
|
|
5GY0
 
 | | Crystal structure of endoglucanase CelQ from Clostridium thermocellum complexed with cellotetraose | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structures of the C-Terminally Truncated Endoglucanase Cel9Q from Clostridium thermocellum Complexed with Cellodextrins and Tris.
Chembiochem, 20, 2019
|
|
3TYK
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | Descriptor: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
5P9T
 
 | | humanized rat catechol O-methyltransferase in complex with 6-(4-fluorophenyl)-8-hydroxy-3H-quinazolin-4-one | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 6-(4-fluorophenyl)-8-oxidanyl-3~{H}-quinazolin-4-one, ... | | Authors: | Ehler, A, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2016-10-19 | | Release date: | 2017-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
7GY9
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 6.84 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
5PBN
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 8) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1YT9
 
 | | HIV Protease with oximinoarylsulfonamide bound | | Descriptor: | (S)-N-((2S,3R)-3-HYDROXY-4-(4-((E)-(HYDROXYIMINO)METHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YL)-3-METHYL-2-(3 -((2-METHYLTHIAZOL-4-YL)METHYL)-2-OXOIMIDAZOLIDIN-1-YL)BUTANAMIDE, Pol polyprotein | | Authors: | Yeung, C.M, Klein, L.L, Flentge, C.A, Randolph, J.T, Zhao, C, Sun, M, Dekhtyar, T, Stoll, V.S, Kempf, D.J. | | Deposit date: | 2005-02-10 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oximinoarylsulfonamides as potent HIV protease inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7GYD
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 11.40 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
5PC2
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 23) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3U1X
 
 | |
5PCJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 40) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PD1
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 57) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PDP
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 82) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PE6
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 99) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7GUO
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 1 at 18.12 MGy X-ray dose. | | Descriptor: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
3DCF
 
 | |
7GY5
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 2.28 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
5PEG
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 109) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7GY8
 
 | | Crystal Structure of HSP72 in complex with ligand 10 at 5.70 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
