8PKR
 
 | |
8PMF
 
 | |
7G1Y
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-methoxyphenyl)sulfanylmethyl]-1,3-thiazole-4-carboxylic acid | | Descriptor: | 2-{[(3-methoxyphenyl)sulfanyl]methyl}-1,3-thiazole-4-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Grenz-Achim, K, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
6TXP
 
 | | Human Aldose Reductase Mutant L300A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid) | | Descriptor: | (2-carbamoyl-5-fluorophenoxy)acetic acid, 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldo-keto reductase family 1 member B1, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Human Aldose Reductase Mutant L300A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid)
To Be Published
|
|
8B66
 
 | |
8GBI
 
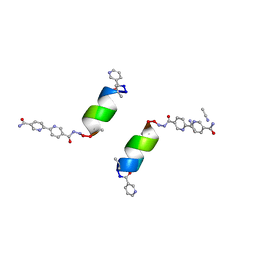 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, LEU-(AIB)-ALA-CYS-LEU-CYS-CYS-(AIB)-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-26 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
5RCV
 
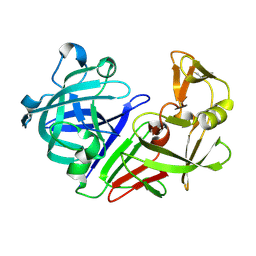 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 17 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1MJ5
 
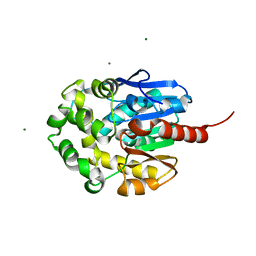 | | LINB (haloalkane dehalogenase) from sphingomonas paucimobilis UT26 at atomic resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Oakley, A.J, Damborsky, J, Wilce, M.C. | | Deposit date: | 2002-08-27 | | Release date: | 2003-08-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 A resolution: dynamics of catalytic residues.
Biochemistry, 43, 2004
|
|
8GKB
 
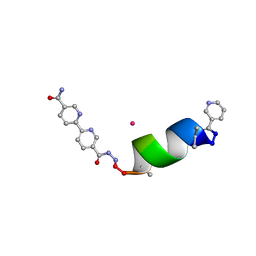 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, CADMIUM ION, LEU-AIB-ALA-SER-LEU-ALA-CYS-AIB-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
3VLA
 
 | | Crystal structure of edgp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EDGP | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
8SY4
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-SER-LEU-ALA-SNC-AIB-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-05-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
5RBW
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library D06b | | Descriptor: | (3-methoxyphenyl)(pyrrolidin-1-yl)methanone, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6TWT
 
 | | Crystal structure of N-terminally truncated NDM-1 metallo-beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Imiolczyk, B, Czyrko-Horczak, J, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
4O6Q
 
 | | 0.95A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (Y75A mutant) | | Descriptor: | FORMIC ACID, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
6YTU
 
 | | Atomic-resolution structure of the coiled-coil dimerisation domain of human Arc | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Activity-regulated cytoskeleton-associated protein, CHLORIDE ION | | Authors: | Hallin, E.I, Touma, C, Bramham, C.R, Kursula, P. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Arc self-association and formation of virus-like capsids are mediated by an N-terminal helical coil motif.
Febs J., 288, 2021
|
|
4WKA
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7LTV
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 3 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LTB
 
 | | Crystal Structure of Keratinicyclin B | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, 3-ammonio-2,3,6-trideoxy-alpha-L-arabino-hexopyranose-(1-2)-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural and Functional Analysis of Keratinicyclin Reveals Synergistic Antibiosis with Vancomycin against Clostridium difficile
to be published
|
|
7LKC
 
 | | Crystal Structure of Keratinimicin A | | Descriptor: | (2~{S},4~{S},5~{R},6~{S})-4-azanyl-5-methoxy-6-methyl-oxan-2-ol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Davis, K.M, Jeffrey, P.D, Seyedsayamdost, M.R. | | Deposit date: | 2021-02-02 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural and Functional Analysis of Keratinicyclin Reveals Synergistic Antibiosis with Vancomycin against Clostridium difficile
to be published
|
|
6UGB
 
 | | C3 symmetric peptide design number 2, Baby Basil | | Descriptor: | C3 symmetric peptide design number 2, Baby Basil, CHLORIDE ION, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
7OV0
 
 | |
7OTU
 
 | | Human OMPD-domain of UMPS in complex with 6-hydroxy-UMP at 0.95 Angstroms resolution, crystal 2 | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Isoform 2 of Uridine 5'-monophosphate synthase | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6RI8
 
 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Third structure of the series with 800 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, FLUORIDE ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1WY3
 
 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH7.0 | | Descriptor: | Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5E9N
 
 | | Steccherinum murashkinskyi laccase at 0.95 resolution | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Glazunova, O.A, Fedorova, T.V, Koroleva, O.V. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi.
Int. J. Biol. Macromol., 118, 2018
|
|
