2XVA
 
 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with sinefungin | | Descriptor: | SINEFUNGIN, TELLURITE RESISTANCE PROTEIN TEHB, ZINC ION | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
4B9D
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
4NMH
 
 | | 11-beta-HSD1 in complex with a 3,3-Di-methyl-azetidin-2-one | | Descriptor: | (4S)-4-(2-methoxyphenyl)-3,3-dimethyl-1-[3-(methylsulfonyl)phenyl]azetidin-2-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McCoull, W, Augustin, M, Blake, C, Ertan, A, Kilgour, E.K, Krapp, S, Moore, J.E, Newcombe, N.J, Packer, M.J, Rees, A, Revill, J, Scott, J.S, Selmi, N, Gerhardt, S, Ogg, D.J, Steinbacher, S, Whittamore, P.R.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and optimisation of 3,3-dimethyl-azetidin-2-ones as potent and selective inhibitors of 11 beta-hydroxysteroid dehydrogenase type 1 (11-beta-HSD1)
TO BE PUBLISHED
|
|
7F58
 
 | | Cryo-EM structure of THIQ-MC4R-Gs_Nb35 complex | | Descriptor: | (3R)-N-[(2R)-3-(4-chlorophenyl)-1-[4-cyclohexyl-4-(1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-1-oxidanylidene-propan-2-yl]-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
2IOG
 
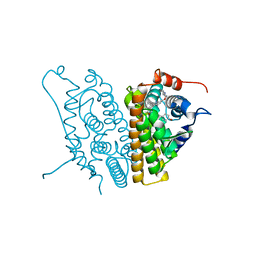 | |
4NMM
 
 | | Crystal Structure of a G12C Oncogenic Variant of Human KRas Bound to a Novel GDP Competitive Covalent Inhibitor | | Descriptor: | 5'-O-[(S)-{[(S)-[2-(acetylamino)ethoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]guanosine, GTPase KRas, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Lim, S.M, Westover, K.D. | | Deposit date: | 2013-11-15 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2VFY
 
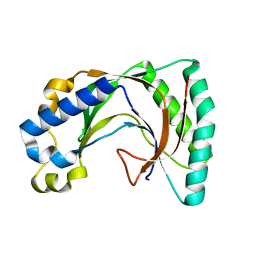 | |
2XDE
 
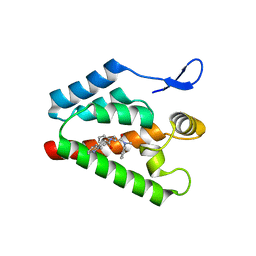 | | Crystal structure of the complex of PF-3450074 with an engineered HIV capsid N terminal domain | | Descriptor: | GAG POLYPROTEIN, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | Authors: | Brown, D.G, Irving, S.L, Anderson, M, Bazin, R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | HIV Capsid is a Tractable Target for Small Molecule Therapeutic Intervention.
Plos Pathog., 6, 2010
|
|
1YJM
 
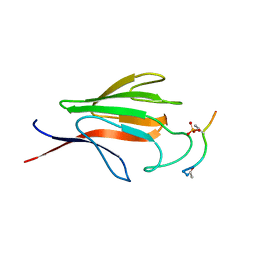 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
3N81
 
 | |
4FKA
 
 | | High resolution structure of the manganese derivative of insulin | | Descriptor: | Insulin A chain, Insulin B chain, MANGANESE (II) ION, ... | | Authors: | Prugovecki, B, Pulic, I, Toth, M, Matkovic-Calogovic, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High Resolution Structure of the Manganese Derivative of Insulin
Croat.Chem.Acta, 85, 2012
|
|
1D7E
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
4M1J
 
 | | Crystal structure of Pseudomonas aeruginosa PvdQ in complex with a transition state analogue | | Descriptor: | Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, GLYCEROL, ... | | Authors: | Wu, R, Clevenger, K, Er, J, Fast, W.L, Liu, D. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of a Transition State Analogue with Picomolar Affinity for Pseudomonas aeruginosa PvdQ, a Siderophore Biosynthetic Enzyme.
Acs Chem.Biol., 8, 2013
|
|
1FZ8
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II COCRYSTALLIZED WITH DIBROMOMETHANE | | Descriptor: | CALCIUM ION, DIBROMOMETHANE, FE (III) ION, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
2IRU
 
 | |
3V53
 
 | | Crystal structure of human RBM25 | | Descriptor: | RNA-binding protein 25 | | Authors: | Gong, D.S. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional characterization of the human RBM25 PWI domain and its flanking basic region
Biochem.J., 450, 2013
|
|
1UHL
 
 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
3NIP
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase complexed with 1,6-diaminohexane | | Descriptor: | 3-guanidinopropionase, HEXANE-1,6-DIAMINE | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
1KYR
 
 | | Crystal Structure of a Cu-bound Green Fluorescent Protein Zn Biosensor | | Descriptor: | COPPER (II) ION, Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
7F53
 
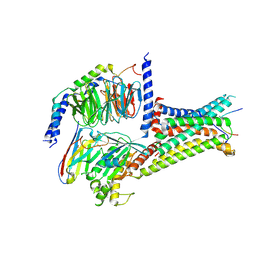 | | Cryo-EM structure of a-MSH-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
4YF0
 
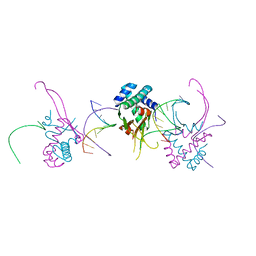 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
3UXK
 
 | |
4GV7
 
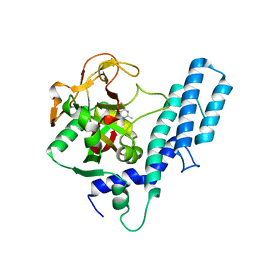 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor ME0328 | | Descriptor: | 2-methylquinazolin-4(3H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | PARP Inhibitor with Selectivity Toward ADP-Ribosyltransferase ARTD3/PARP3
Acs Chem.Biol., 8, 2013
|
|
2NN1
 
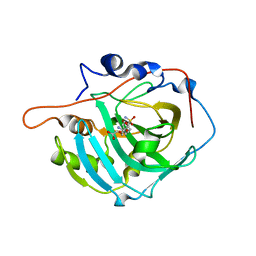 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 1, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
4H7C
 
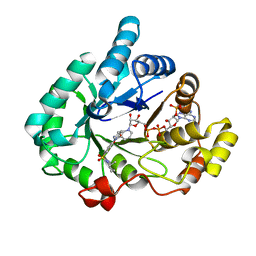 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 1-{4-[(2-methyl-1-piperidinyl)sulfonyl]phenyl}-2-pyrrolidinone | | Descriptor: | 1-(4-{[(2R)-2-methylpiperidin-1-yl]sulfonyl}phenyl)-1,3-dihydro-2H-pyrrol-2-one, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Turnbull, A.P, Heinrich, D, Jamieson, S.M.F, Flanagan, J.U, Silva, S, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Denny, W.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Synthesis and structure-activity relationships for 1-(4-(piperidin-1-ylsulfonyl)phenyl)pyrrolidin-2-ones as novel non-carboxylate inhibitors of the aldo-keto reductase enzyme AKR1C3.
Eur.J.Med.Chem., 62C, 2013
|
|
