3HO7
 
 | |
3LS1
 
 | | Crystal Structure of Cyanobacterial PsbQ from Synechocystis sp. PCC 6803 complexed with Zn2+ | | Descriptor: | Sll1638 protein, ZINC ION | | Authors: | Jackson, S.A, Fagerlund, R.D, Wilbanks, S.M, Eaton-Rye, J.J. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 A: Implications for Binding and Function in Cyanobacterial Photosystem II
Biochemistry, 49, 2010
|
|
3LS6
 
 | | Crystal structure of 3,4-Dihydroxy-2-butanone 4-phosphate synthase in complex with sulfate and zinc | | Descriptor: | 3,4-Dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumar, P, Karthikeyan, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Potential anti-bacterial drug target: structural characterization of 3,4-dihydroxy-2-butanone-4-phosphate synthase from Salmonella typhimurium LT2.
Proteins, 78, 2010
|
|
3LC2
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from methicillin resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-09 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3LEF
 
 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1Z6NA_001 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein 4E10_S0_1Z6NA_001 (T18) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-14 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3T0M
 
 | |
3LL0
 
 | | Monomeric Griffithsin with two Gly-Ser Insertions | | Descriptor: | GLYCEROL, Griffithsin, SULFATE ION | | Authors: | Moulaei, T, Wlodawer, A. | | Deposit date: | 2010-01-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity.
Structure, 18, 2010
|
|
3LF6
 
 | | Crystal structure of HIV epitope-scaffold 4E10_1XIZA_S0_001_N | | Descriptor: | PHOSPHATE ION, Putative phosphotransferase system | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LFO
 
 | |
3LKM
 
 | |
3Q89
 
 | |
3Q8U
 
 | |
3KSZ
 
 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD and G3P | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-11-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
4Q0L
 
 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
3TQD
 
 | | Structure of the 3-deoxy-D-manno-octulosonate cytidylyltransferase (kdsB) from Coxiella burnetii | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase, ACETATE ION, NICKEL (II) ION | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3LC1
 
 | | Crystal Structure of H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.0 angstrom resolution. | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3L6O
 
 | | Crystal Structure of Phosphate bound apo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.2 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, PHOSPHATE ION | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-12-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3S34
 
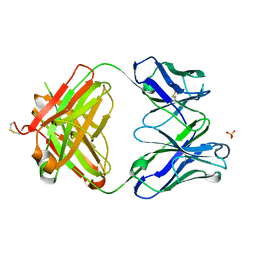 | | Structure of the 1121B Fab fragment | | Descriptor: | 1121B Fab heavy chain, 1121B Fab light chain, PHOSPHATE ION | | Authors: | Franklin, M.C. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Function of Two Anti-VEGF Receptor 2 Antibodies.
Structure, 19, 2011
|
|
3S6L
 
 | |
3LF9
 
 | | Crystal structure of HIV epitope-scaffold 4E10_D0_1IS1A_001_C | | Descriptor: | 4E10_D0_1IS1A_001_C (T161) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-16 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LX8
 
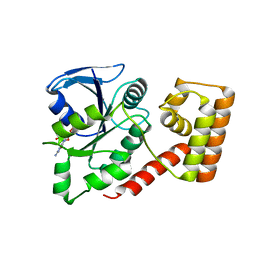 | | Crystal structure of GDP-bound NFeoB from S. thermophilus | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
3LVF
 
 | |
3U04
 
 | |
4RMF
 
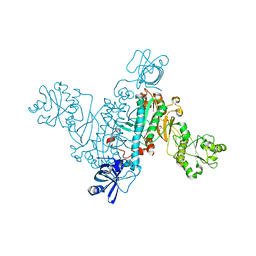 | | Biochemical and structural characterization of mycobacterial aspartyl-tRNA synthetase AspS, a promising TB drug target | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Aspartate--tRNA(Asp/Asn) ligase, FORMIC ACID | | Authors: | Cox, J.A.G, Gurcha, S.S, Veeraraghavan, U, Besra, G.S, Futterer, K. | | Deposit date: | 2014-10-21 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Characterization of Mycobacterial Aspartyl-tRNA Synthetase AspS, a Promising TB Drug Target.
Plos One, 9, 2014
|
|
3UCL
 
 | |
