5HUA
 
 | | Structure of C. glabrata FKBP12-FK506 complex | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-01-27 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Pathogenic Fungal FKBP12s Reveal Possible Self-Catalysis Function.
Mbio, 7, 2016
|
|
5HKG
 
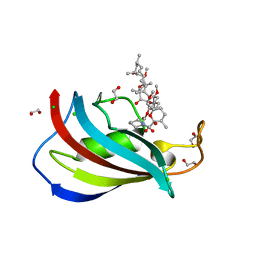 | | Total chemical synthesis, refolding and crystallographic structure of a fully active immunophilin: calstabin 2 (FKBP12.6). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Sirigu, S, Huet, T, Bacchi, M, Jullian, M, Fould, B, Ferry, G, Vuillard, L, Chavas, L, Puget, K, Nosjean, O, Boutin, J.A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Total chemical synthesis, refolding, and crystallographic structure of fully active immunophilin calstabin 2 (FKBP12.6).
Protein Sci., 25, 2016
|
|
7UJ5
 
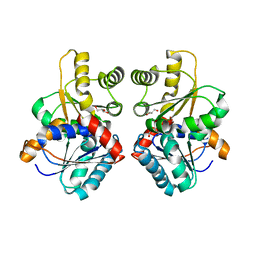 | |
1A33
 
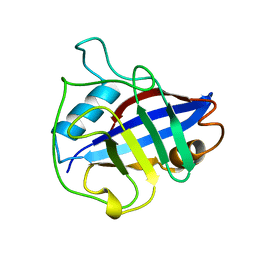 | |
5ZR0
 
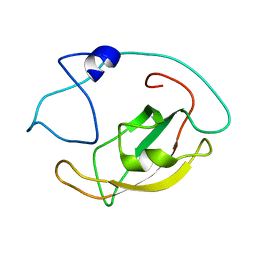 | | Solution structure of peptidyl-prolyl cis/trans isomerase domain of Trigger Factor in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein,Trigger factor | | Authors: | Kawagoe, S, Nakagawa, H, Kumeta, H, Ishimori, K, Saio, T. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into prolinecis/transisomerization of unfolded proteins catalyzed by the trigger factor chaperone.
J. Biol. Chem., 293, 2018
|
|
5HTG
 
 | |
1B1A
 
 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Hoffmann, B, Konrat, R, Bothe, H, Buckel, W, Kraeutler, B. | | Deposit date: | 1998-11-19 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium.
Eur.J.Biochem., 263, 1999
|
|
7ZEY
 
 | | Complex Cyp33-RRM : MLL1-PHD3 | | Descriptor: | MLL cleavage product N320, Peptidyl-prolyl cis-trans isomerase E, ZINC ION | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
5IJW
 
 | | Glutamate Racemase (MurI) from Mycobacterium smegmatis with bound D-glutamate, 1.8 Angstrom resolution, X-ray diffraction | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase, IODIDE ION | | Authors: | Poen, S, Nakatani, Y, Krause, K. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery.
Biochem. J., 473, 2016
|
|
5ZG5
 
 | | Crystal Structure of Triosephosphate isomerase SADsubAAA mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
1AWS
 
 | | SECYPA COMPLEXED WITH HAGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWQ
 
 | | CYPA COMPLEXED WITH HAGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1BE1
 
 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Tollinger, M, Konrat, R, Hilbert, B.H, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum
Structure, 6, 1998
|
|
1AWU
 
 | | CYPA COMPLEXED WITH HVGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
4EM6
 
 | |
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
5BY2
 
 | |
4ELZ
 
 | | CCDBVFI:GYRA14VFI | | Descriptor: | CcdB, DNA gyrase subunit A, GLYCEROL | | Authors: | De Jonge, N, Simic, M, Buts, L, Haesaerts, S, Roelants, K, Garcia-Pino, A, Sterckx, Y, De Greve, H, Lah, J, Loris, R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative interactions define gyrase specificity in the CcdB family.
Mol.Microbiol., 84, 2012
|
|
4EKZ
 
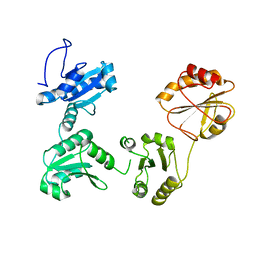 | | Crystal structure of reduced hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4EYV
 
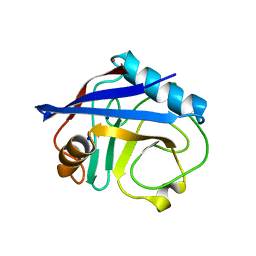 | | Crystal structure of Cyclophilin A like protein from Piriformospora indica | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase, ... | | Authors: | Bhatt, H, Pal, R.K, Tuteja, N, Bhavesh, N.S. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of RNA-interacting Cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants
Sci Rep, 3, 2013
|
|
4GJI
 
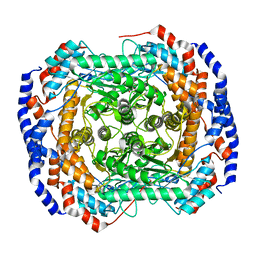 | |
4GIA
 
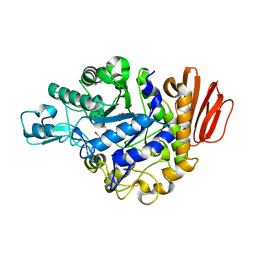 | | Crystal structure of the MUTB F164L mutant from crystals soaked with isomaltulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GI9
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with Trehalulose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7JMH
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 35 - State 4 (S4) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
4GI8
 
 | | Crystal structure of the MUTB F164L mutant from crystals soaked with the substrate sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-08 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity
Acta Crystallogr.,Sect.D, 69, 2013
|
|
