7EA7
 
 | | crystal structure of NAP1 LIR in complex with GABARAP | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, NAP1_LIR motif | | Authors: | Fu, T, Pan, L. | | Deposit date: | 2021-03-06 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural and biochemical advances on the recruitment of the autophagy-initiating ULK and TBK1 complexes by autophagy receptor NDP52.
Sci Adv, 7, 2021
|
|
2QDJ
 
 | | Crystal structure of the Retinoblastoma protein N-domain provides insight into tumor suppression, ligand interaction and holoprotein architecture | | Descriptor: | Retinoblastoma-associated protein | | Authors: | Hassler, M, Mittnacht, S, Pearl, L.H. | | Deposit date: | 2007-06-21 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture.
Mol.Cell, 28, 2007
|
|
6Y09
 
 | |
6ZAY
 
 | | Crystal structure of Atg16L in complex with GDP-bound Rab33B | | Descriptor: | 1,2-ETHANEDIOL, Autophagy-related protein 16-1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pantoom, S, Wu, Y.W. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | RAB33B recruits the ATG16L1 complex to the phagophore via a noncanonical RAB binding protein.
Autophagy, 17, 2021
|
|
3POM
 
 | |
5XUY
 
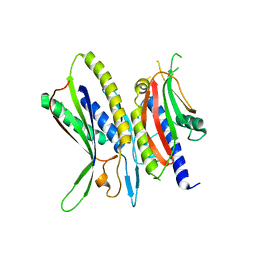 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV6
 
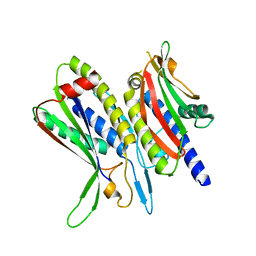 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV4
 
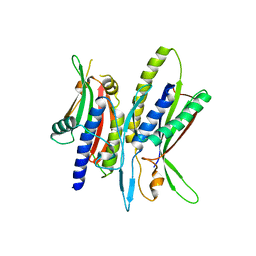 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV3
 
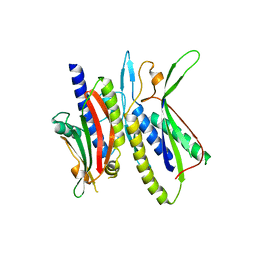 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13, DI(HYDROXYETHYL)ETHER | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
2AZE
 
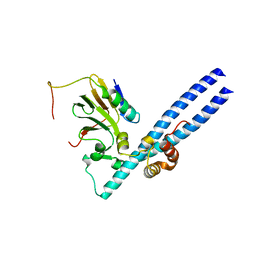 | | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer | | Descriptor: | Retinoblastoma-associated protein, Transcription factor Dp-1, Transcription factor E2F1 | | Authors: | Rubin, S.M, Gall, A.L, Zheng, N, Pavletich, N.P. | | Deposit date: | 2005-09-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release.
Cell(Cambridge,Mass.), 123, 2005
|
|
2R7G
 
 | |
1RKD
 
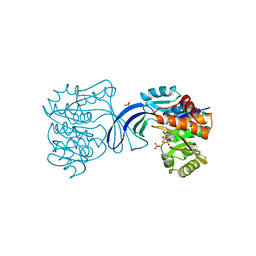 | | E. COLI RIBOKINASE COMPLEXED WITH RIBOSE AND ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOKINASE, ... | | Authors: | Sigrell, J.A, Cameron, A.D, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1997-11-29 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 A resolution: insights into a new family of kinase structures.
Structure, 6, 1998
|
|
1RKS
 
 | |
4ELL
 
 | |
4ELJ
 
 | |
5Z9S
 
 | |
7O3V
 
 | | Stalk complex structure (TrwJ/VirB5-TrwI/VirB6) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwI protein, TrwJ protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O41
 
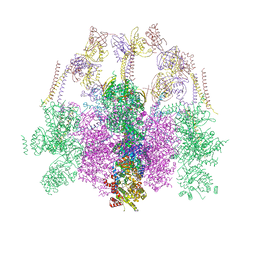 | | Hexameric composite model of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
6OJX
 
 | |
6OKV
 
 | |
6OLM
 
 | |
6OLK
 
 | |
6OJZ
 
 | |
7OIU
 
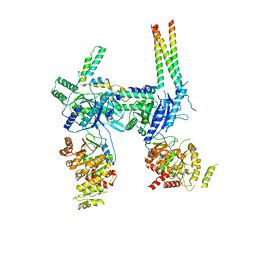 | | Inner Membrane Complex (IMC) protomer structure (TrwM/VirB3, TrwK/VirB4, TrwG/VirB8tails) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-05-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7Q1V
 
 | |
