4JZE
 
 | | Structure of factor VIIA in complex with the inhibitor 2-{2-[(1-aminoisoquinolin-6-yl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid | | Descriptor: | 2-{2-[(1-aminoisoquinolin-6-yl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Wei, A, Anumula, R. | | Deposit date: | 2013-04-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of nonbenzamidine factor VIIa inhibitors using a biaryl acid scaffold.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JZD
 
 | | Structure of factor VIIA in complex with the inhibitor 2-{2-[(4-carbamimidoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid | | Descriptor: | 2-{2-[(4-carbamimidoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Jacobson, B.L, Anumula, R. | | Deposit date: | 2013-04-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of nonbenzamidine factor VIIa inhibitors using a biaryl acid scaffold.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3M7R
 
 | | Crystal structure of VDR H305Q mutant | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Vitamin D3 receptor | | Authors: | Rochel, N, Hourai, S, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
6F8X
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-26g | | Descriptor: | 1,2-ETHANEDIOL, 2-[(5~{R})-3-(3-cyclopentyloxy-4-methoxy-phenyl)-4,5-dihydro-1,2-oxazol-5-yl]-~{N},~{N}-bis(2-hydroxyethyl)ethanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F9T
 
 | | Crystal structure of human testis Angiotensin-1 converting enzyme in complex with Sampatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of sampatrilat and sampatrilat-Asp in complex with human ACE - a molecular basis for domain selectivity.
FEBS J., 285, 2018
|
|
1MVQ
 
 | | Cratylia mollis lectin (isoform 1) in complex with methyl-alpha-D-mannose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin, ... | | Authors: | de Souza, G.A, Oliveira, P.S, Trapani, S, Correia, M.T, Oliva, G, Coelho, L.C, Greene, L.J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Amino acid sequence and tertiary structure of Cratylia mollis seed lectin
Glycobiology, 13, 2003
|
|
5TKK
 
 | |
1SN9
 
 | |
1SNA
 
 | | An Oligomeric Domain-Swapped Beta-Beta-Alpha Mini-Protein | | Descriptor: | ISOPROPYL ALCOHOL, tetrameric beta-beta-alpha mini-protein | | Authors: | Ali, M.H, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SNE
 
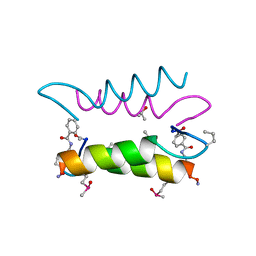 | | An Oligomeric Domain-Swapped Beta-Beta-Alpha Mini-Protein | | Descriptor: | ISOPROPYL ALCOHOL, tetrameric beta-beta-alpha mini-protein | | Authors: | Ali, M.H, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5A8O
 
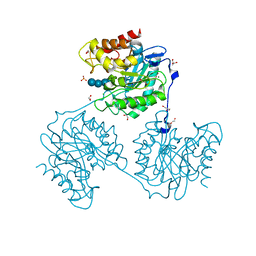 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with cellotetraose | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
6EZ4
 
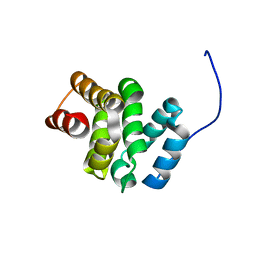 | | NMR structure of the C-terminal domain of the human RPAP3 protein | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Fabre, P, Chagot, M.E, Bragantini, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones.
Nat Commun, 9, 2018
|
|
6MS3
 
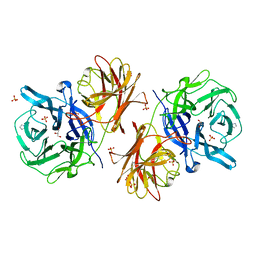 | | Crystal structure of the GH43 protein BlXynB mutant (K247S) from Bacillus licheniformis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside Hydrolase Family 43, ... | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
6Q6M
 
 | |
5A94
 
 | | Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PUTATIVE RETAINING B-GLYCOSIDASE, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
6MOB
 
 | | Crystal structure of KIT1 in complex with DP2976 via co-crystallization | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-{4-chloro-5-[1-ethyl-7-(methylamino)-2-oxo-1,2-dihydro-1,6-naphthyridin-3-yl]-2-fluorophenyl}-N'-phenylurea, NITRATE ION | | Authors: | Edwards, T.E, Abendroth, J, Safford, K, Chun, L. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ripretinib (DCC-2618) Is a Switch Control Kinase Inhibitor of a Broad Spectrum of Oncogenic and Drug-Resistant KIT and PDGFRA Variants.
Cancer Cell, 35, 2019
|
|
6MRM
 
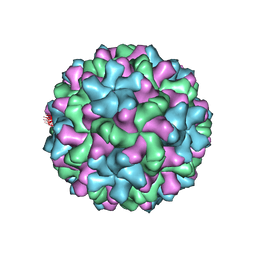 | | Red Clover Necrotic Mosaic Virus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
6MWE
 
 | | CRYSTAL STRUCTURE OF TIE2 IN COMPLEX WITH DECIPERA COMPOUND DP1919 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Angiopoietin-1 receptor, SULFATE ION | | Authors: | Chun, L, Abendroth, J, Edwards, T.E. | | Deposit date: | 2018-10-29 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Selective Tie2 Inhibitor Rebastinib Blocks Recruitment and Function of Tie2
Mol. Cancer Ther., 16, 2017
|
|
2B7C
 
 | | Yeast guanine nucleotide exchange factor eEF1Balpha K205A mutant in complex with eEF1A | | Descriptor: | Elongation factor 1-alpha, elongation factor-1 beta | | Authors: | Pittman, Y.R, Valente, L, Jeppesen, M.G, Andersen, G.R, Patel, S, Kinzy, T.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha
J.Biol.Chem., 281, 2006
|
|
5A8M
 
 | | Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
5AA7
 
 | | Structural and functional characterization of a chitin-active 15.5 kDa lytic polysaccharide monooxygenase domain from a modular chitinase from Jonesia denitrificans | | Descriptor: | CHITINASE, COPPER (I) ION | | Authors: | Mekasha, S, Forsberg, Z, Dalhus, B, Choudhary, S, Schmidt-Dannert, C, Vaaje-Kolstad, G, Eijsink, V. | | Deposit date: | 2015-07-23 | | Release date: | 2015-12-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of a Small Chitin-Active Lytic Polysaccharide Monooxygenase Domain of a Multi-Modular Chitinase from Jonesia Denitrificans.
FEBS Lett., 590, 2016
|
|
6GDA
 
 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P43212 form soaked with Spermine | | Descriptor: | Cytochrome c iso-1, HEME C, SPERMINE, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tuning Protein Frameworks via Auxiliary Supramolecular Interactions.
Acs Nano, 13, 2019
|
|
1FXU
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CALF SPLEEN IN COMPLEX WITH N(7)-ACYCLOGUANOSINE INHIBITOR AND A PHOSPHATE ION | | Descriptor: | 2-AMINO-7-[2-(2-HYDROXY-1-HYDROXYMETHYL-ETHYLAMINO)-ETHYL]-1,7-DIHYDRO-PURIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Luic, M, Bzowska, A, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase: structure of its ternary complex with an N(7)-acycloguanosine inhibitor and a phosphate anion.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5A95
 
 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
4WQP
 
 | |
