4PXY
 
 | |
4Q7P
 
 | |
4Q87
 
 | |
3B5E
 
 | |
4LF1
 
 | | Hexameric Form II RuBisCO from Rhodopseudomonas palustris, activated and complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Chan, S, Satagopan, S, Sawaya, M.R, Eisenberg, D, Tabita, F.R, Perry, L.J. | | Deposit date: | 2013-06-26 | | Release date: | 2014-06-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris.
J.Biol.Chem., 289, 2014
|
|
4HNI
 
 | | crystal structure of ck1e in complex with PF4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform epsilon, SULFATE ION | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
7KEP
 
 | | avibactam-CDD-1 2 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
4HW6
 
 | |
4D4A
 
 | | Structure of the catalytic domain (BcGH76) of the Bacillus circulans GH76 alpha mannanase, Aman6. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3C86
 
 | | OpdA from agrobacterium radiobacter with bound product diethyl thiophosphate from crystal soaking with tetraethyl dithiopyrophosphate- 1.8 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2008-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
4IAJ
 
 | |
6MOM
 
 | | Crystal structure of human Interleukin-1 receptor associated Kinase 4 (IRAK 4, CID 100300) in complex with compound NCC00371481 (BSI 107591) | | Descriptor: | 1,2-ETHANEDIOL, 6-[7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-3-yl]-N-[(3R)-pyrrolidin-3-yl]pyridin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Abendroth, J, Mayclin, S.J, Lorimer, D.D, Starczynowski, D, Hoyt, S, Tawa, G, Thomas, C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Overcoming adaptive therapy resistance in AML by targeting immune response pathways.
Sci Transl Med, 11, 2019
|
|
1NPW
 
 | | Crystal structure of HIV protease complexed with LGZ479 | | Descriptor: | CARBAMIC ACID 1-{5-BENZYL-5-[2-HYDROXY-4-PHENYL-3-(TETRAHYDRO-FURAN- 3-YLOXYCARBONYLAMINO)-BUTYL]-4-OXO-4,5-DIHYDRO-1H-PYRROL-3-YL}- INDAN-2-YL ESTER, POL polyprotein | | Authors: | Smith III, A.B. | | Deposit date: | 2003-01-20 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors.
J.Med.Chem., 46, 2003
|
|
4LGN
 
 | | The structure of Acidothermus cellulolyticus family 74 glycoside hydrolase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellulose-binding, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Acidothermus cellulolyticus family 74 glycoside hydrolase at 1.82 angstrom resolution.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ZA7
 
 | | Structure of A. niger Fdc1 in complex with alpha-methyl cinnamic acid | | Descriptor: | (2E)-2-methyl-3-phenylprop-2-enoic acid, 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4H5F
 
 | | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1
TO BE PUBLISHED
|
|
3R90
 
 | | Crystal structure of Malignant T cell-amplified sequence 1 protein | | Descriptor: | GLYCEROL, Malignant T cell-amplified sequence 1, SULFATE ION, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Tong, Y, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Malignant T cell-amplified sequence 1 protein
to be published
|
|
6BTL
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD117 1-[2-(Piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
5ET5
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4O81
 
 | |
3WBG
 
 | | Structure of the human heart fatty acid-binding protein in complex with 1-anilinonaphtalene-8-sulphonic acid | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Fatty acid-binding protein, heart | | Authors: | Hirose, M, Sugiyama, S, Ishida, H, Niiyama, M, Matsuoka, D, Hara, T, Sato, F, Mizohata, E, Murakami, S, Inoue, T, Matsuoka, S, Murata, M. | | Deposit date: | 2013-05-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the human-heart fatty-acid-binding protein 3 in complex with the fluorescent probe 1-anilinonaphthalene-8-sulphonic acid
J.SYNCHROTRON RADIAT., 20, 2013
|
|
5ZGQ
 
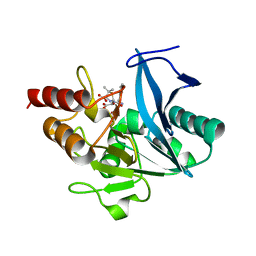 | | Crystal structure of NDM-1 at pH7.5 (Tris-HCl, (NH4)2SO4) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGE
 
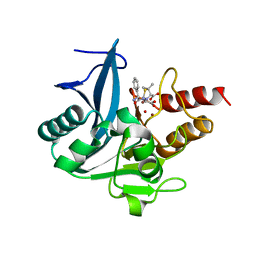 | | Crystal structure of NDM-1 at pH5.5 (Bis-Tris) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGR
 
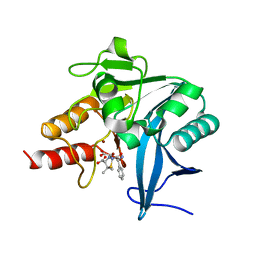 | | Crystal structure of NDM-1 at pH7.3 (HEPES) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-10 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
4Q53
 
 | | Crystal structure of a DUF4783 family protein (BACUNI_04292) from Bacteroides uniformis ATCC 8492 at 1.27 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CHLORIDE ION, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of a hypothetical protein (BACUNI_04292) from Bacteroides uniformis ATCC 8492 at 1.27 A resolution
To be published
|
|
