7E4A
 
 | | Crystal structure of MIF bound to compound 13 | | Descriptor: | CHLORIDE ION, Flavoxate, Macrophage migration inhibitory factor, ... | | Authors: | Fa, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
3IPE
 
 | | Human Transthyretin (TTR) complexed with a palindromic bivalent amyloid inhibitor (7 carbon linker). | | Descriptor: | 2,2'-{heptane-1,7-diylbis[oxy(3,5-dichlorobenzene-4,1-diyl)imino]}dibenzoic acid, Transthyretin | | Authors: | Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Trapping of palindromic ligands within native transthyretin prevents amyloid formation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7E49
 
 | | Crystal structure of MIF bound to compound10 | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.574 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
4CEA
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (R)-[2-[[(2R)-butan-2-yl]carbamoyl]phenyl]methyl-[[(2S)-5-carboxy-2-(2-carboxyethyl)-2,3-dihydro-1,4-benzodioxin-6-yl]methyl]-prop-2-enyl-azanium, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
2A57
 
 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
9HY3
 
 | | sc-4E (scFv derived from the mAb 4E1 against CD93) crystallized at pH 5.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural and antigen-binding surface definition of an anti-CD93 monoclonal antibody for the treatment of degenerative vascular eye diseases.
Int.J.Biol.Macromol., 309, 2025
|
|
4QJG
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin V
To be Published
|
|
2WKS
 
 | | Structure of Helicobacter pylori Type II Dehydroquinase with a new carbasugar-thiophene inhibitor. | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Guardado-Calvo, P, Llamas-Saiz, A.L, Prazeres, V.F.V, Tizon, L, Castedo, L, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and biological evaluation of new nanomolar competitive inhibitors of Helicobacter pylori type II dehydroquinase. Structural details of the role of the aromatic moieties with essential residues.
J. Med. Chem., 53, 2010
|
|
4QK2
 
 | |
5NWE
 
 | | Complex of H275Y mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
4QMN
 
 | | MST3 in complex with BOSUTINIB | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Serine/threonine-protein kinase 24 | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
5SHN
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(cnn1C)C(NCc2ncco2)=O)C(Nc4nc3nc(cn3cc4)c5ccccc5)=O, micromolar IC50=0.016936 | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SI6
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c4(c(C(Nc2cc1nc(cn1cc2C#N)c3ccccc3)=O)n(C)nc4)C(=O)N5CCC5, micromolar IC50=0.025368 | | Descriptor: | 4-(azetidine-1-carbonyl)-N-[(4R)-6-cyano-2-phenylimidazo[1,2-a]pyridin-7-yl]-1-methyl-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SEL
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c3(c(C(Nc1cc(nn1C)c2ncccc2)=O)n(nc3)C)C(=O)N4CCC4, micromolar IC50=0.00796 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[1-methyl-3-(pyridin-2-yl)-1H-pyrazol-5-yl]-1H-pyrazole-5-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
2WKM
 
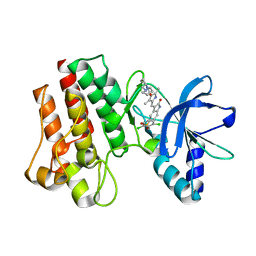 | | X-ray Structure of PHA-00665752 bound to the kinase domain of c-Met | | Descriptor: | (3Z)-5-[(2,6-DICHLOROBENZYL)SULFONYL]-3-[(3,5-DIMETHYL-4-{[(2S)-2-(PYRROLIDIN-1-YLMETHYL)PYRROLIDIN-1-YL]CARBONYL}-1H-PYRROL-2-YL)METHYLIDENE]-1,3-DIHYDRO-2H-INDOL-2-ONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2009-06-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Drug Design of Crizotinib (Pf-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal-Epithelial Transition Factor (C-met) Kinase and Anaplastic Lymphoma Kinase (Alk).
J.Med.Chem, 54, 2011
|
|
6HXK
 
 | |
9HY1
 
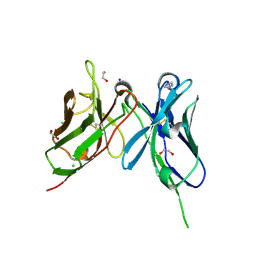 | | sc-4E (scFv derived from the mAb 4E1 against CD93) crystallized at pH 7.5 (dimeric state from gel filtration) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and antigen-binding surface definition of an anti-CD93 monoclonal antibody for the treatment of degenerative vascular eye diseases.
Int.J.Biol.Macromol., 309, 2025
|
|
6KTX
 
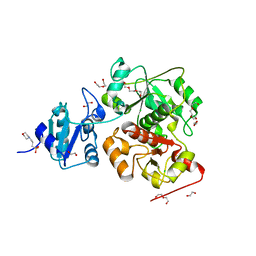 | | The wildtype structure of EanB | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
4PY5
 
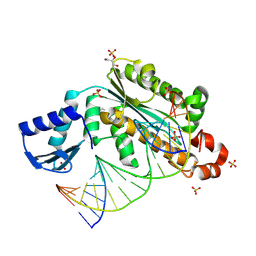 | | Thermovibrio ammonificans RNase H3 in complex with 19-mer RNA/DNA | | Descriptor: | 5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*TP*C)-3', 5'-R(*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3', GLYCEROL, ... | | Authors: | Figiel, M, Nowotny, M. | | Deposit date: | 2014-03-26 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition.
Nucleic Acids Res., 42, 2014
|
|
4Q4M
 
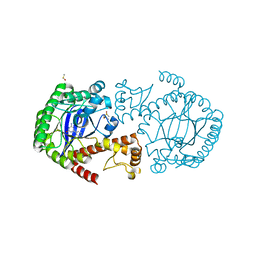 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 6-Amino-4-phenyl-1,2-dihydro-1,3,5-triazin-2-one | | Descriptor: | 6-amino-4-phenyl-1,3,5-triazin-2(1H)-one, DIMETHYL SULFOXIDE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | 5-Azacytosines as a Novel Scaffold to Inhibit Z. mobilis TGT with Expected Improved Bioavailability and Synthetic Accessibility
To be Published
|
|
1DWJ
 
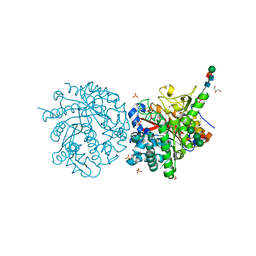 | |
5C3Q
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and thymine (T) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, NICKEL (II) ION, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
5SF8
 
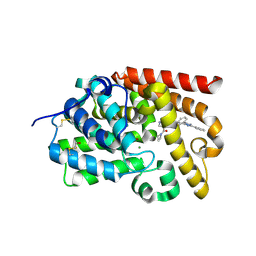 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(cccc1)c2nn(c(n2)CCNC(c3c(cnn3C)C(=O)N4CCC4)=O)c5ccccn5, micromolar IC50=0.000474 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-{2-[3-phenyl-1-(pyridin-2-yl)-1H-1,2,4-triazol-5-yl]ethyl}-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
2WAX
 
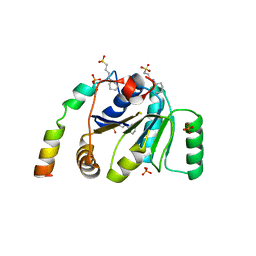 | |
