4C7T
 
 | | Focal Adhesion Kinase catalytic domain in complex with a diarylamino- 1,3,5-triazine inhibitor | | Descriptor: | FOCAL ADHESION KINASE 1, N-methyl-2-[[4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl]amino]benzamide, SULFATE ION | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2013-09-25 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Inhibition of Both Focal Adhesion Kinase and Fibroblast Growth Factor Receptor 2 Pathways Induces Anti-Tumor and Anti-Angiogenic Activities.
Cancer Lett., 348, 2014
|
|
6KQ1
 
 | | Crystal structure of cytochrome c551 from Pseudomonas sp. strain MT-1. | | Descriptor: | Cytochrome C biogenesis protein CcsA, HEME C, ZINC ION | | Authors: | Fujii, S, Oki, H, Kawahara, K, Ohkubo, T, Masanari-Fujii, M, Wakai, S, Sambongi, Y. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into high stability of cytochrome c551 from a deep-sea piezo-tolerant bacterium, Pseudomonas sp. strain MT-1
To Be Published
|
|
4Q99
 
 | |
5SFO
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(cc(c1ccccc1)nc2CCNC(c4c(C(=O)N3CCC3)cnn4C)=O)C, micromolar IC50=0.004013 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
2A3M
 
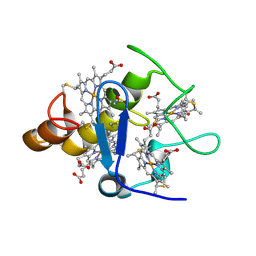 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome (oxidized form) | | Descriptor: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C | | Authors: | Pattarkine, M.V, Tanner, J.J, Bottoms, C.A, Lee, Y.H, Wall, J.D. | | Deposit date: | 2005-06-25 | | Release date: | 2006-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
4CEC
 
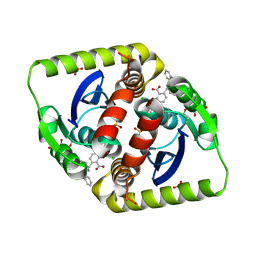 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-6-[[[2-(cyclobutylmethylcarbamoyl)phenyl]methyl-prop-2-enyl-amino]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
5BV8
 
 | | G1324S mutation in von Willebrand Factor A1 domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, von Willebrand factor | | Authors: | Campbell, J.C, Kim, C, Tischer, A, Auton, M. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mutational Constraints on Local Unfolding Inhibit the Rheological Adaptation of von Willebrand Factor.
J.Biol.Chem., 291, 2016
|
|
4CFA
 
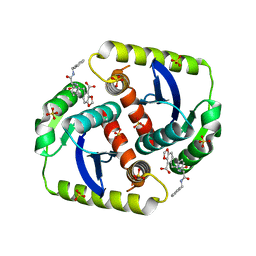 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 5-[[methyl-[[(1S)-2-oxidanylidene-1-phenyl-2-[(phenylmethyl)amino]ethyl]carbamoyl]amino]methyl]-1,3-benzodioxole-4-carboxylic acid, INTEGRASE, SULFATE ION | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4BVW
 
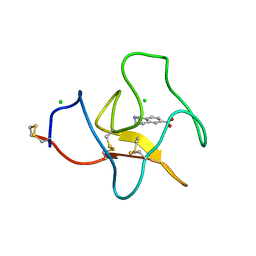 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1,2,3,4-tetrahydroisoquinoline-6-carboxylic acid, 1,2-ETHANEDIOL, APOLIPOPROTEIN(A), ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
2X1D
 
 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, ACETATE ION, ACYL-COENZYME, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
5SJT
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c4(c(C(Nc2cc1nc(cn1cc2)c3ccccc3)=O)n(nc4)C)C(N(C)C)=O, micromolar IC50=0.000153 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4BYI
 
 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-((1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)methyl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
6AG9
 
 | | Crystal structure of uPA in complex with 3,5-bis(azanyl)-6-(1-benzofuran-2-yl)-N-carbamimidoyl-pyrazine-2- carboxamide | | Descriptor: | 3,5-bis(azanyl)-6-(1-benzofuran-2-yl)-N-carbamimidoyl-pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Majed, H, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | uPA-HMA
To Be Published
|
|
6EJZ
 
 | | Tryptophan Repressor TrpR from E.coli variant S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, SULFATE ION, ... | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-24 | | Release date: | 2019-02-06 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
5SFN
 
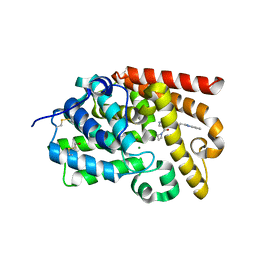 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c12c(cccc1)n(C)c(n2)CCNC(c4c(C(=O)N3CCC3)cnn4C)=O, micromolar IC50=0.01602 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[2-(1-methyl-1H-benzimidazol-2-yl)ethyl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7EL5
 
 | |
1NCC
 
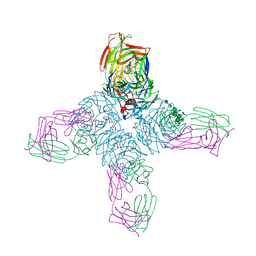 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
4XX3
 
 | |
2AAC
 
 | |
4QG8
 
 | | crystal structure of PKM2-K305Q mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
237L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
5SJE
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C2(=NN(c1cccc(c1)[S](=O)(=O)C)C=CC2=O)c3ccnn3c4cccc5c4OC(O5)(F)F, micromolar IC50=0.029598 | | Descriptor: | 3-[1-(2,2-difluoro-2H-1,3-benzodioxol-4-yl)-1H-pyrazol-5-yl]-1-[3-(methanesulfonyl)phenyl]pyridazin-4(1H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SE7
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(c(c(C(=O)N1CCC1)cn2)C(Nc4cc3nc(nn3cc4)c5ccccc5)=O)C, micromolar IC50=0.00054035 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
4CED
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-6-[[[2-(furan-2-ylmethylcarbamoyl)phenyl]methyl-prop-2-enyl-amino]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CEQ
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-6-[[[2-(cyclohexylmethylcarbamoyl)phenyl]methyl-methyl-amino]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
