7CDD
 
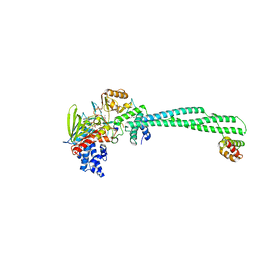 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDE
 
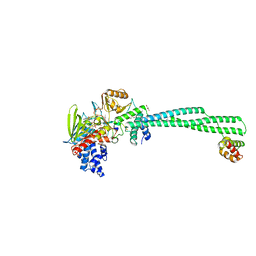 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRKR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
4I36
 
 | | Crystal Structure of the Bacillus stearothermophilus Phosphofructokinase Mutant D12A | | Descriptor: | 6-phosphofructokinase | | Authors: | Mosser, R, Reddy, M, Bruning, J.B, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2012-11-25 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redefining the Role of the Quaternary Shift in Bacillus stearothermophilus Phosphofructokinase.
Biochemistry, 52, 2013
|
|
6E0S
 
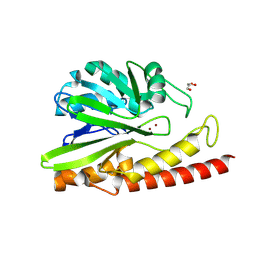 | | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library | | Descriptor: | GLYCEROL, MEM-A1, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Lau, C, Topp, E, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library
To Be Published
|
|
6E8A
 
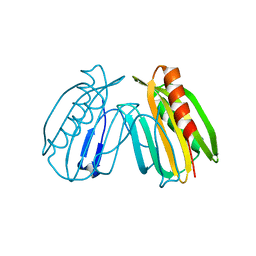 | | Crystal structure of DcrB from Salmonella enterica at 1.92 Angstroms resolution | | Descriptor: | DUF1795 domain-containing protein | | Authors: | Rasmussen, D.M, Soens, R.W, Bhattacharyya, B, May, J.F. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of DcrB, a lipoprotein from Salmonella enterica, reveals flexibility in the N-terminal segment of the Mog1p/PsbP-like fold.
J. Struct. Biol., 204, 2018
|
|
6EN6
 
 | | Crystal structure B of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
4OHM
 
 | | Human GKRP bound to AMG-0771 and sorbitol-6-phosphate | | Descriptor: | (2S)-2-{4-[(2S)-4-[(6-aminopyridin-3-yl)sulfonyl]-2-(prop-1-yn-1-yl)piperazin-1-yl]phenyl}-3,3,3-trifluoropropane-1,2-diol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
3LM1
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with p-nitrophenyl-GalNAc | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
4XI0
 
 | | MamA 41-end from Desulfovibrio magneticus RS-1 | | Descriptor: | Magnetosome protein MamA | | Authors: | Zarivach, R, Zeytuni, N, Cronin, S, Davidov, G, Baran, D, Stein, T. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MamA as a Model Protein for Structure-Based Insight into the Evolutionary Origins of Magnetotactic Bacteria.
Plos One, 10, 2015
|
|
6KZL
 
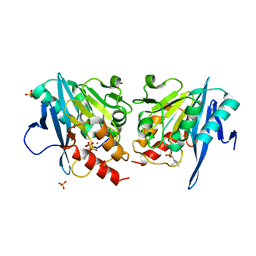 | | Crystal Structure Of NDM-1 Metallo-beta-lactamase In Complex With Inhibitor X2 | | Descriptor: | 2,5-diethyl-1-methyl-4-sulfamoyl-pyrrole-3-carboxylic acid, Beta-lactamase, SULFATE ION, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
5U5N
 
 | |
6KXO
 
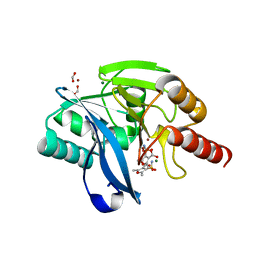 | | Crystal Structure Of VIM-2 Metallo-beta-lactamase In Complex With Inhibitor NO9 | | Descriptor: | 2,5-dimethyl-4-sulfamoyl-furan-3-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
3LVW
 
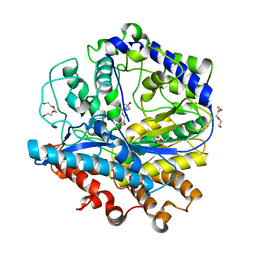 | | Glutathione-inhibited ScGCL | | Descriptor: | GLUTATHIONE, Glutamate--cysteine ligase, TRIETHYLENE GLYCOL | | Authors: | Biterova, E.I, Barycki, J.J. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for feedback and pharmacological inhibition of Saccharomyces cerevisiae glutamate cysteine ligase.
J.Biol.Chem., 285, 2010
|
|
6KTS
 
 | | Structure of C34N126K/N36 | | Descriptor: | Envelope glycoprotein, Glycoprotein 41 | | Authors: | Yu, D.W, Qin, B. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Characterization of the Secondary Mutation N126K Selected by Various HIV-1 Fusion Inhibitors.
Viruses, 12, 2020
|
|
6KUF
 
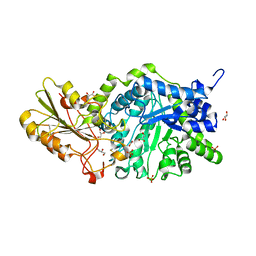 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with glucose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-09-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
4J2R
 
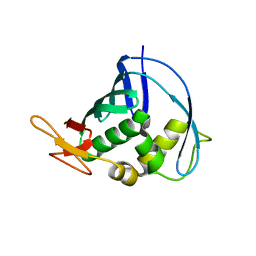 | | Middle domain of influenza A virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Qiu, H, Tsurumura, T, Tsumori, Y, Hatakeyama, D, Kuzuhara, T, Tsuge, H. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of a surface mutant of the middle domain of PB2 from human influenza A (H1N1) virus
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
6L1J
 
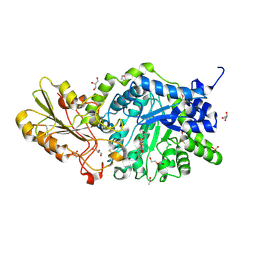 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4'-nitrophenyl thiolaminaritrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
5TYT
 
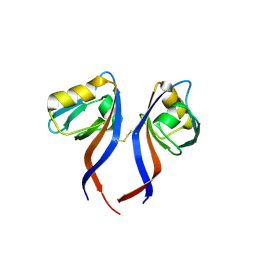 | | Crystal Structure of the PDZ domain of RhoGEF bound to CXCR2 C-terminal peptide | | Descriptor: | Rho guanine nucleotide exchange factor 11, C-X-C chemokine receptor type 2 chimera | | Authors: | Spellmon, N, Holcomb, J, Niu, A, Choudhary, V, Sun, X, Brunzelle, J, Li, C, Yang, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF.
Biochem. Biophys. Res. Commun., 485, 2017
|
|
5UFR
 
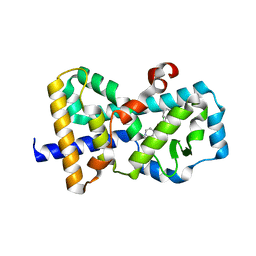 | | Structure of RORgt bound to | | Descriptor: | (S)-[4-chloro-2-(dimethylamino)-3-phenylquinolin-6-yl](1-methyl-1H-imidazol-5-yl)(pyridin-4-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7E0F
 
 | |
5UA0
 
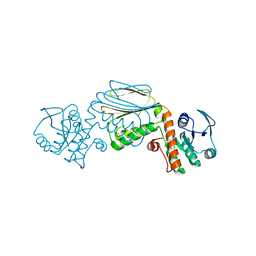 | |
6ED9
 
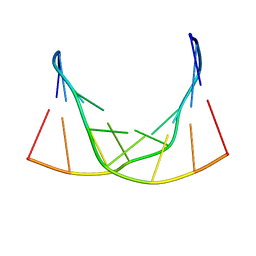 | |
3LLZ
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with Gal-beta-1,3-GalNAc | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
6EN5
 
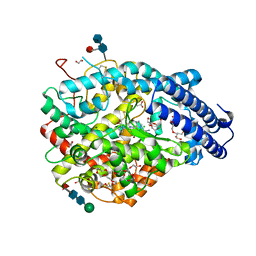 | | Crystal structure A of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
4JB8
 
 | | Caspase-7 in Complex with DARPin C7_16 | | Descriptor: | Caspase-7 subunit p11, Caspase-7 subunit p20, DARPin C7_16 | | Authors: | Fluetsch, A, Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
