5ZAJ
 
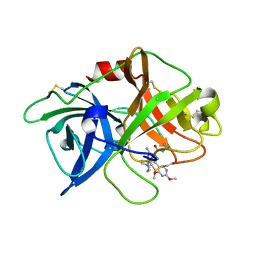 | | uPA-31F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(2,4-dimethoxypyrimidin-5-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator chain B | | Authors: | Buckley, B.J, Jiang, L.G, Huang, M.D, Kelso, M.J, Ranson, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 6-Substituted Hexamethylene Amiloride (HMA) Derivatives as Potent and Selective Inhibitors of the Human Urokinase Plasminogen Activator for Use in Cancer.
J. Med. Chem., 61, 2018
|
|
5VT8
 
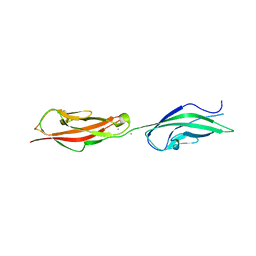 | |
8FP5
 
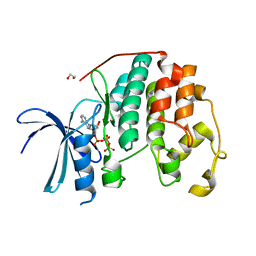 | | CDK2 liganded with ATP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclin-dependent kinase 2, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
5W1D
 
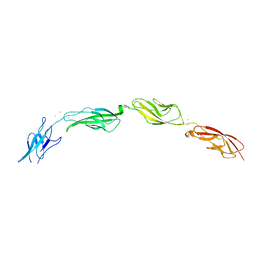 | |
6K2R
 
 | |
6DTK
 
 | | Heterodimers of FALS mutant SOD enzyme | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase C111S/D83S-C111S HETERODIMER, ... | | Authors: | Streltsov, V.A, Nuttall, S.D, Ganio, K.E, Roberts, B. | | Deposit date: | 2018-06-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of heterodimers of FALS mutant SOD enzyme
To Be Published
|
|
6KH8
 
 | | Solution structure of Zn free Bovine Pancreatic Insulin in 20% acetic acid-d4 (pH 1.9) | | Descriptor: | Insulin A Chain, Insulin B chain | | Authors: | Bhunia, A, Ratha, B.N, Kar, R.K, Brender, J.R. | | Deposit date: | 2019-07-14 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-18 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
5W4T
 
 | | Crystal Structure of Fish Cadherin-23 EC1-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
7LS3
 
 | | Co-complex CYP46A1 with 8114 (3f) | | Descriptor: | (5-methyl-2-pyridin-4-yl-phenyl)-[4-oxidanyl-4-(phenylmethyl)piperidin-1-yl]methanone, Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-02-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Soticlestat, a Potent and Selective Inhibitor for Cholesterol 24-Hydroxylase (CH24H).
J.Med.Chem., 64, 2021
|
|
5YXZ
 
 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | Authors: | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484
To Be Published
|
|
7TND
 
 | |
5YY1
 
 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-[2-(trifluoromethyl)phenyl]quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | Authors: | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739
To Be Published
|
|
5VZQ
 
 | | Sperm whale myoglobin V68A/I107Y with nitric oxide | | Descriptor: | Myoglobin, NITRIC OXIDE, NITRITE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
7TNF
 
 | |
7LRL
 
 | | Co-complex CYP46A1 with 7742 (Soticlestat/TAK-935)) | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, [4-oxidanyl-4-(phenylmethyl)piperidin-1-yl]-(2-pyridin-4-ylpyridin-3-yl)methanone | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-02-16 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Discovery of Soticlestat, a Potent and Selective Inhibitor for Cholesterol 24-Hydroxylase (CH24H).
J.Med.Chem., 64, 2021
|
|
7TNU
 
 | |
6D52
 
 | |
7LS4
 
 | | Co-complex CYP46A1 with 9129 (1b) | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, [5,5-dimethyl-3-(2-methylphenyl)-4~{H}-pyrazol-1-yl]-pyridin-4-yl-methanone | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-02-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Soticlestat, a Potent and Selective Inhibitor for Cholesterol 24-Hydroxylase (CH24H).
J.Med.Chem., 64, 2021
|
|
7TP6
 
 | |
7TP5
 
 | |
4P9V
 
 | | Grb2 SH2 complexed with a pTyr-Ac6cN-Asn tripeptide | | Descriptor: | CHLORIDE ION, Growth factor receptor-bound protein 2, PHQ-PTR-02K-ASN-NH2 | | Authors: | Clements, J.H, Martin, S.F. | | Deposit date: | 2014-04-06 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Protein-ligand interactions: Probing the energetics of a putative cation-pi interaction.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7TQM
 
 | |
5W26
 
 | | INFLUENZA VIRUS NEURAMINIDASE N9 IN COMPLEX WITH 4-DEOXYGENATED 2,3-DIFLUORO-N-ACETYLNEURAMINIC ACID | | Descriptor: | (2R,3R,5R,6R)-5-acetamido-2,3-bis(fluoranyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (2~{R},3~{R},5~{R})-3-acetamido-5-fluoranyl-2-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]-2,3,4,5-tetrahydropyran-1-ium-6-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Mckimm-Breschkin, J, Barrett, S, Pilling, P, Hader, S, Watt, A.G. | | Deposit date: | 2017-06-05 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Anti-Influenza Activity of 4-, 7-, 8- and 9-Deoxygenated 2,3-Difluoro- N-acetylneuraminic Acid Derivatives.
J. Med. Chem., 61, 2018
|
|
5VP7
 
 | | Crystal structure of human KRAS G12A mutant in complex with GDP | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, S, Long, B, Boris, G, Chen, A, Ni, S, Kennedy, M.A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4P2P
 
 | | AN INDEPENDENT CRYSTALLOGRAPHIC REFINEMENT OF PORCINE PHOSPHOLIPASE A2 AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Finzel, B.C, Ohlendorf, D.H, Weber, P.C, Salemme, F.R. | | Deposit date: | 1991-10-22 | | Release date: | 1992-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An independent crystallographic refinement of porcine phospholipase A2 at 2.4 A resolution
Acta Crystallogr.,Sect.B, 47, 1991
|
|
