9GV7
 
 | | Structure of reverse docking TCR in complex with peptide-HLA | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Karuppiah, V. | | Deposit date: | 2024-09-23 | | Release date: | 2025-04-23 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Determining T-cell receptor binding orientation and Peptide-HLA interactions using cross-linking mass spectrometry.
J.Biol.Chem., 301, 2025
|
|
4Z09
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Thr2040Ala] peptide | | Descriptor: | Apical membrane antigen 1, GLYCEROL, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4C1P
 
 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | Descriptor: | BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | Deposit date: | 2013-08-13 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C2N
 
 | | Crystal structure of human testis angiotensin-I converting enzyme mutant E403R | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Yates, C.J, Schwager, S.L.U, Mohd, A, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2013-08-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular and Thermodynamic Mechanisms of the Chloride Dependent Human Angiotensin-I Converting Enzyme (Ace)
J.Biol.Chem., 289, 2014
|
|
7K8Z
 
 | |
4RJD
 
 | | TFP bound in alternate orientations to calcium-saturated Calmodulin C-Domains | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Feldkamp, M.D, Gakhar, L, Pandey, N, Shea, M.A. | | Deposit date: | 2014-10-08 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Opposing orientations of the anti-psychotic drug trifluoperazine selected by alternate conformations of M144 in calmodulin.
Proteins, 83, 2015
|
|
5CT3
 
 | | The structure of the NK1 fragment of HGF/SF complexed with 2FA | | Descriptor: | 3-hydroxypropane-1-sulfonic acid, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5O7W
 
 | | Crystal structure of the 4-FLUORO RSL lectin in complex with Lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative fucose-binding lectin protein, ... | | Authors: | Varrot, A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Effect of Noncanonical Amino Acids on Protein-Carbohydrate Interactions: Structure, Dynamics, and Carbohydrate Affinity of a Lectin Engineered with Fluorinated Tryptophan Analogs.
ACS Chem. Biol., 13, 2018
|
|
5WAD
 
 | | ADC-7 in complex with boronic acid transition state inhibitor CR161 | | Descriptor: | Beta-lactamase, PHOSPHATE ION, phosphonooxy-[[[6-(1~{H}-1,2,3,4-tetrazol-5-yl)pyridin-3-yl]sulfonylamino]methyl]borinic acid | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
4Z9A
 
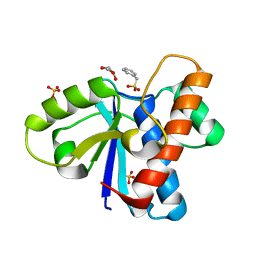 | | Crystal structure of Low Molecular Weight Protein Tyrosine Phosphatase isoform A complexed with phenylmethanesulfonic acid | | Descriptor: | GLYCEROL, Low molecular weight phosphotyrosine protein phosphatase, SULFATE ION, ... | | Authors: | Trivella, D.B.B, Fonseca, E.M.B, Scorsato, V, Dias, M.P, Aparicio, R. | | Deposit date: | 2015-04-10 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the apo form and a complex of human LMW-PTP with a phosphonic acid provide new evidence of a secondary site potentially related to the anchorage of natural substrates.
Bioorg.Med.Chem., 23, 2015
|
|
3GHM
 
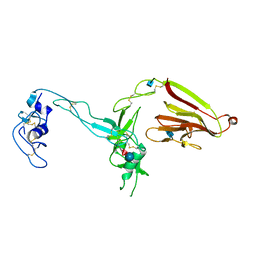 | | Crystal structure of the exosite-containing fragment of human ADAMTS13 (form-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A disintegrin and metalloproteinase with thrombospondin motifs 13, ... | | Authors: | Akiyama, M, Takeda, S, Kokame, K, Takagi, J, Miyata, T. | | Deposit date: | 2009-03-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the non-catalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor
To be Published
|
|
9J5X
 
 | | Cryo-EM Structure of URAT1 in Complex with Lingolinurad | | Descriptor: | 3-bromanyl-5-(2-ethylimidazo[1,2-a]pyridin-3-yl)carbonyl-2-oxidanyl-benzenecarbonitrile, Solute carrier family 22 member 12 | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2024-08-13 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Basis for Inhibition of Urate Reabsorption in URAT1.
Jacs Au, 5, 2025
|
|
5WF5
 
 | | Agonist bound human A2a adenosine receptor with D52N mutation at 2.60 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Human A2a adenosine receptor T4L chimera | | Authors: | White, K.L, Eddy, M.T, Gao, Z, Han, G.W, Hanson, M.A, Lian, T, Deary, A, Patel, N, Jacobson, K.A, Katritch, V, Stevens, R.C. | | Deposit date: | 2017-07-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling.
Structure, 26, 2018
|
|
5SAA
 
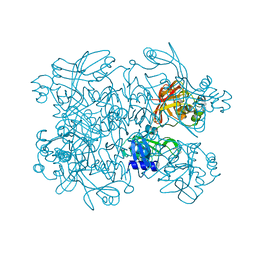 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z319891284 | | Descriptor: | 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.239 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAD
 
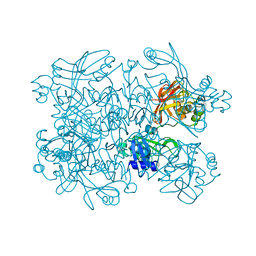 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z425449682 | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
2AXY
 
 | | Crystal Structure of KH1 domain of human Poly(C)-binding protein-2 with C-rich strand of human telomeric DNA | | Descriptor: | C-rich strand of human telomeric dna, Poly(rC)-binding protein 2 | | Authors: | Du, Z, Lee, J.K, Tjhen, R.J, Li, S, Stroud, R.M, James, T.L. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the First KH Domain of Human Poly(C)-binding Protein-2 in Complex with a C-rich Strand of Human Telomeric DNA at 1.7 A
J.Biol.Chem., 280, 2005
|
|
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
7EXZ
 
 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
4AW0
 
 | | Human PDK1 Kinase Domain in Complex with Allosteric Compound PS182 Bound to the PIF-Pocket | | Descriptor: | 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Schulze, J.O, Busschots, K, Lopez-Garcia, L.A, Lammi, C, Stroba, A, Zeuzem, S, Piiper, A, Alzari, P.M, Neimanis, S, Arencibia, J.M, Engel, M, Biondi, R.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Substrate-Selective Inhibition of Protein Kinase Pdk1 by Small Compounds that Bind to the Pif-Pocket Allosteric Docking Site.
Chem.Biol., 19, 2012
|
|
6E0E
 
 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
5S84
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010949a | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4R)-1,3-oxazolidin-4-yl]methanamine, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8B
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010960a | | Descriptor: | 1,2-ETHANEDIOL, 1lambda~6~-thietane-1,1-dione, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5AZB
 
 | | Crystal structure of Escherichia coli Lgt in complex with phosphatidylglycerol and the inhibitor palmitic acid | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PALMITIC ACID, Prolipoprotein diacylglyceryl transferase, ... | | Authors: | Zhang, X.C, Mao, G, Zhao, Y. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E. coli lipoprotein diacylglyceryl transferase
Nat Commun, 7, 2016
|
|
6BXS
 
 | |
3KEH
 
 | | Crystal Structure of N370S Glucocerebrosidase mutant at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, R.R, Boucher, S, Pan, C.Q, Edmunds, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glucocerebrosidase Containing the N370S mutation: Implication on Chaperon Therapy
To be Published
|
|
